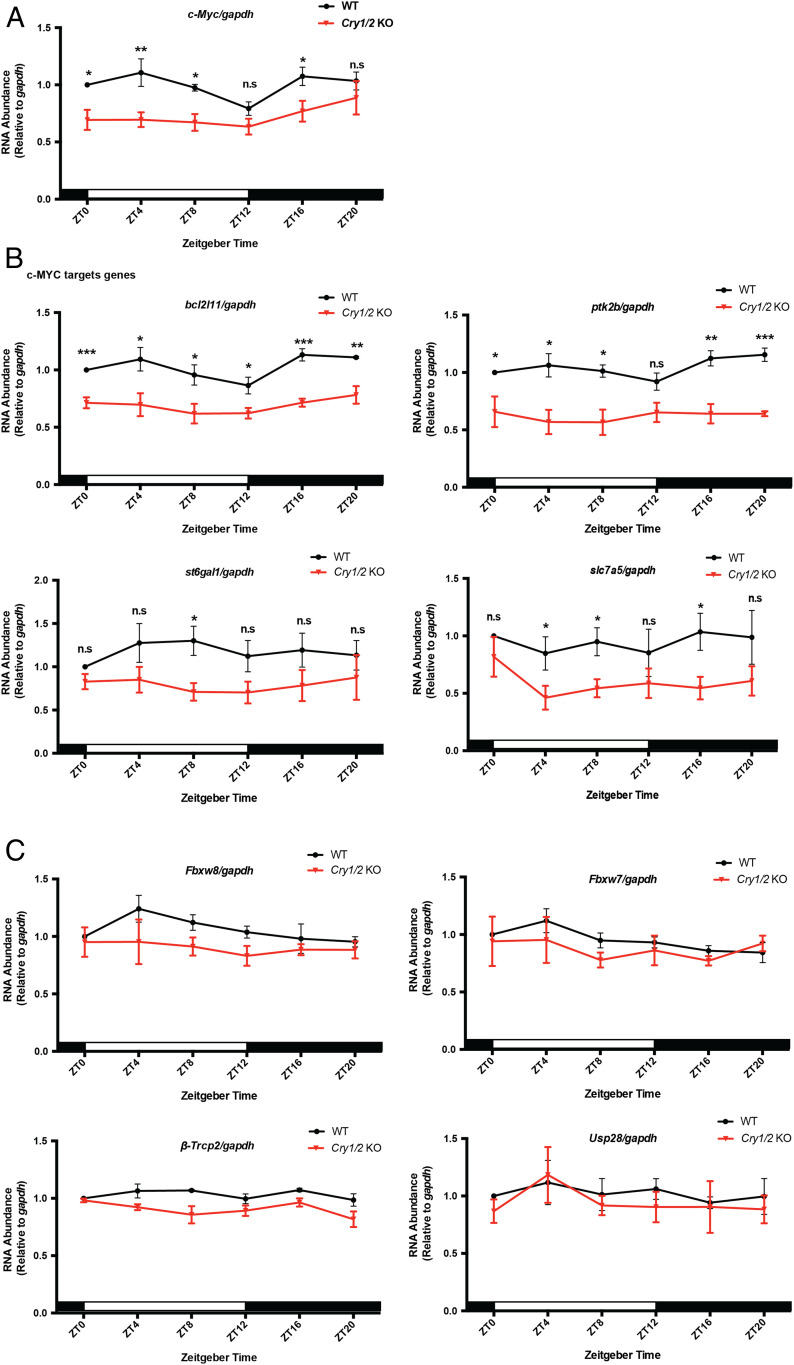
Fig. 4.
CRYs modulate c-MYC expression through transcriptional regulation. (A) c-Myc levels in the spleen samples of WT and Cry1/2 KO mice detected by reverse transcription-qPCR. Three biological repeats were used for quantification. White and black bars indicate lights on and lights off, respectively. (B) mRNA levels of c-MYC target genes in the spleens of WT and Cry1/2 KO. For each genotype and time point, four mice were used for quantification. (C) Expression of genes that regulate c-MYC degradation were measured by reverse transcription-qPCR in spleens of WT and Cry1/2 KO mice using gapdh as an internal control. Samples were collected at the indicated time points. Error bars correspond to SEM. Data were normalized to a value of 1 for WT at ZT0. n.s, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, as determined by the t test.