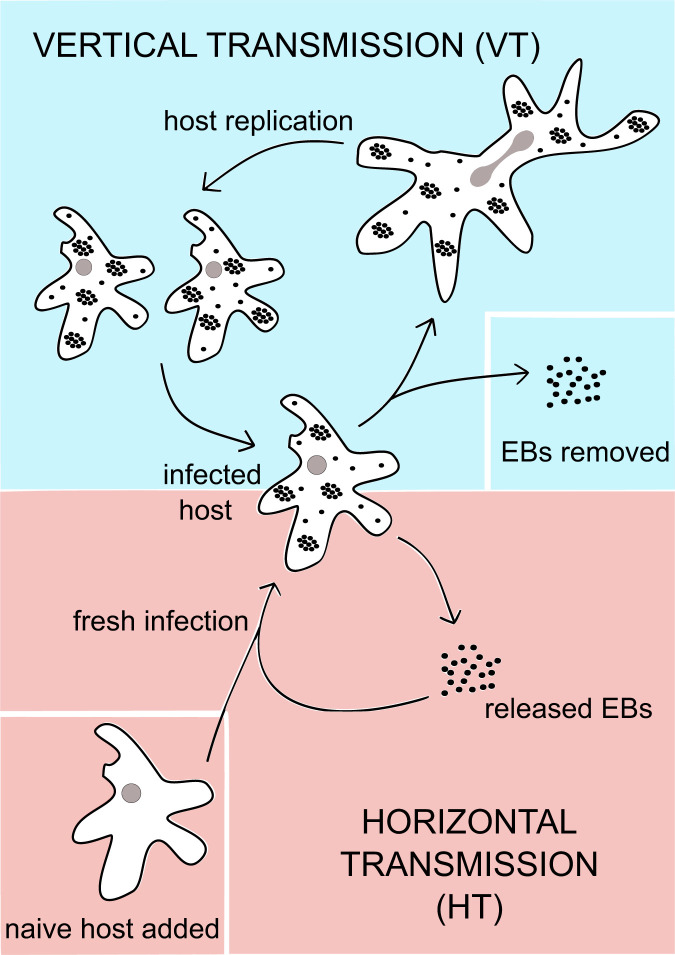
Fig. 1.
The Parachlamydia/Acanthamoeba model and the design of the evolution experiment. Two separate selection regimes were established to select for HT and VT, both originating from the same initial ancestral amoeba–symbiont population. In VT (Top), amoebae that were fully infected with the symbiont were maintained in liquid culture. Daily removal of the liquid medium was carried out to remove released EBs, minimizing the potential for HT. No new amoebae were added to the cultures, and the bacterial symbionts were passed on from parent cell to daughter cell. This was performed for 14 mo, equivalent to 525 symbiont generations. In HT (Bottom), EBs were allowed to accumulate over a week in the liquid medium, after which they were isolated and used to infect naive host cells. Infected amoebae were discarded at the end of the week. Fresh infections, by way of HT, were required for the symbionts to be maintained in the population. This was performed for 14 mo, equivalent to 560 symbiont generations.