Significance
Enhancers are crucial regulatory elements that dictate cellular processes by spatiotemporally controlling promoter activity. Recent studies have shown that active enhancers themselves generate noncoding enhancer RNAs (eRNAs) that play a critical role in fine tuning gene expression. Using genomic approaches, we demonstrate that oncogenic KSHV modulates host enhancer activity in PELs to control its lifecycle. During KSHV latency, expression levels of several key transcription factors and oncogenes are elevated by clusters of enhancers. Upon virus reactivation, global host enhancer activities are suppressed in order to facilitate viral replication. Our results indicate that better understanding and targeting host enhancer and eRNA functions may lead to novel therapeutics for PELs.
Keywords: Kaposi’s sarcoma-associated virus, primary effusion lymphoma, viral epigenetics, viral genomics, enhancer
Abstract
Enhancers play indispensable roles in cell proliferation and survival through spatiotemporally regulating gene transcription. Active enhancers and superenhancers often produce noncoding enhancer RNAs (eRNAs) that precisely control RNA polymerase II activity. Kaposi’s sarcoma-associated herpesvirus (KSHV) is a human oncogenic gamma-2 herpesvirus that causes Kaposi’s sarcoma and primary effusion lymphoma (PEL). It is well characterized that KSHV utilizes host epigenetic machineries to control the switch between two lifecycles, latency and lytic replication. However, how KSHV impacts host epigenome at different stages of viral lifecycle is not well understood. Using global run-on sequencing (GRO-seq) and chromatin-immunoprecipitation sequencing (ChIP-seq), we profiled the dynamics of host transcriptional regulatory elements during latency and lytic replication of KSHV-infected PEL cells. This revealed that a number of critical host genes for KSHV latency, including MYC proto-oncogene, were under the control of superenhancers whose activities were globally repressed upon viral reactivation. The eRNA-expressing MYC superenhancers were located downstream of the MYC gene in KSHV-infected PELs and played a key role in MYC expression. RNAi-mediated depletion or dCas9-KRAB CRISPR inhibition of eRNA expression significantly reduced MYC mRNA level in PELs, as did the treatment of an epigenomic drug that globally blocks superenhancer function. Finally, while cellular IRF4 acted upon eRNA expression and superenhancer function for MYC expression during latency, KSHV viral IRF4 repressed cellular IRF4 expression, decreasing MYC expression and thereby, facilitating lytic replication. These results indicate that KSHV acts as an epigenomic driver that modifies host epigenomic status upon reactivation by effectively regulating host enhancer function.
Enhancers are major distal regulatory elements that control gene expression by recruiting transcription machineries, and are essential for cell type-specific biological processes (1). Superenhancers are clusters of enhancers across a long range of genomic DNA marked by high level of histone H3 lysine 27 acetylation (H3K27ac) and histone H3 lysine 4 monomethylation (H3K4me1) (2). There is considerable evidence indicating that superenhancers are present near genes essential for cell survival and tumorigenesis in order to contribute to the oncogenic process by activating neighboring genes (3, 4). These superenhancers often produce noncoding transcripts called enhancer RNAs (eRNAs) that have been shown recently to play functional roles in enhancer activity, potentially by facilitating promoter–enhancer interactions and recruiting transcription machinery (5, 6).
Herpesviruses have large DNA genomes that replicate in the nucleus and utilize cellular epigenetic processes to control their lifecycles (7). During the default latent state, the viral genome chromatinizes and circularizes and recruits numerous cellular regulatory factors onto its own genome. Upon changes in host environment, the virus reactivates and enters lytic replication (7–9). Kaposi’s sarcoma-associated herpesvirus (KSHV) is one of the gamma-2 herpesviruses that has been extensively studied because of its ability to cause cancers in immunocompromised patients (10). KSHV is the etiologic agent of Kaposi’s sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman’s disease (MCD) (11). PEL cells depend heavily on KSHV genes for survival and often are coinfected with gamma-1 herpesvirus and Epstein-Barr virus (EBV) (12, 13). In PEL, KSHV maintains a latent status in B cells, where only a small subset of genes that are related to cell proliferation, immune evasion, antiapoptosis, and viral episome maintenance are expressed (14). However, there is a small population of KSHV-infected cells that undergoes spontaneous reactivation in which the rest of the viral genes are expressed in a temporal manner of immediate early (IE), early (E), and late (L) genes—eventually leading to production of viral progeny (15). While the majority of KSHV-associated cancers harbor the latent viral genome, this spontaneous reactivation is necessary in order to maintain viral load and contribute to oncogenesis (16). An IE gene ORF50 encodes RTA (replication and transcription activator) which is sufficient and necessary to drive the lytic cycle (17, 18). Lytic replication also leads to an extensive shutoff of host gene expressions, mainly facilitated by the KSHV ORF37 (viral exonuclease) which promotes global mRNA turnover (19, 20). This ultimately allows the virus to prevent host immune responses and to hijack host transcriptional machinery for its own gene expression and viral replication.
Epigenetic modifications of the KSHV genome is crucial for the switch from latency to lytic replication. Upon initial infection, the KSHV genome circularizes and is subjected to H3K27me3 repressive histone modification along with active H3K4me3 modification, resulting in bivalent chromatin formation that allows rapid viral expressions upon stimulus (21–23). While the epigenetic changes of the viral genome upon reactivation are well understood, how the host epigenome is regulated at different stages of the viral lifecycle is not clear. There are several KSHV-encoded gene products such as RTA and latent protein LANA (latency-associated nuclear antigen) that bind to host cell chromatin to change host gene expression (24, 25). As these studies have focused on individual genes, the understanding of KSHV-mediated host epigenomic changes at a global scale remains unanswered.
Deregulation of c-MYC (MYC) is a hallmark of many lymphomas and other types of cancers (26). Unlike other B cell lymphomas, PEL cells lack major genetic translocations of proto-oncogenes such as MYC (27). On the other hand, KSHV still maintains elevated MYC expression during latency, which is crucial in maintaining latent status and cell survival (28). Several viral latent proteins such as LANA and vIRF3 (viral IFN regulatory factor-3) have been shown to stabilize MYC at the posttranslational level (29, 30). However, whether KSHV regulates MYC gene expression at the transcriptional level has not been well studied. Recently, it has been shown that in EBV-transformed lymphoblastoid cell lines (LCLs), EBV proteins such as EBNA2 and EBNA3C modify host genome by binding to cellular enhancer sites and up-regulating oncogene expressions such as MYC and BCL2, ultimately inducing LCL proliferation (31). Also, the eRNA transcripts produced from the MYC superenhancer regions are critical for maintaining MYC mRNA expression and cell proliferation, suggesting the significant role of eRNAs for EBV pathogenesis (32).
Here, we present a molecular mechanism of how KSHV acts as an epigenomic driver that modulates the host epigenome at different stages of its viral lifecycle in order to maximize successful replication and latency. By applying a combination of global run-on sequencing (GRO-seq), chromatin-immunoprecipitation sequencing (ChIP-seq), and chromosome conformation capture (3C) combined with high-throughput sequencing (4C-seq), we present a comprehensive transcriptomic and epigenomic landscape of KSHV-infected PEL cells during latency and lytic replication. We further characterize superenhancer regulatory elements and eRNAs that control critical genes such as the MYC gene. This study provides detailed insight into the underlying regulatory mechanisms involved in the KSHV lifecycle and lymphomagenesis.
Results
Global Reduction of Host Enhancer and Superenhancer Activity upon KSHV Reactivation.
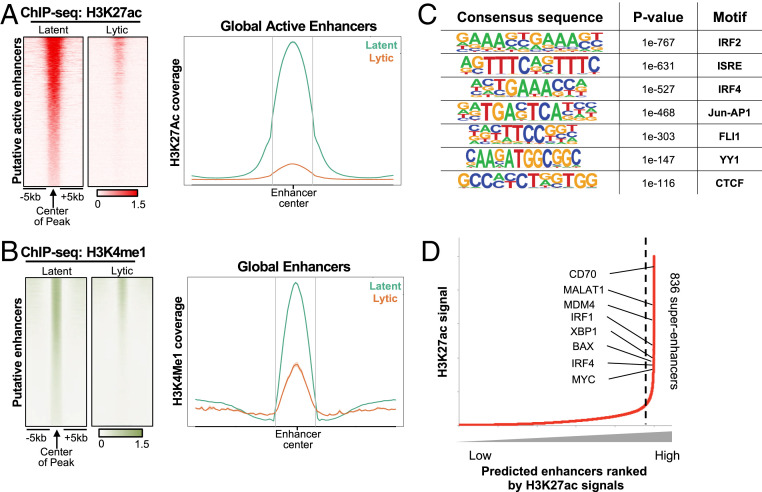
Active enhancers can be identified by the high enrichments of both H3K4me1 and H3K27ac relative to trimethylated H3K4 (H3K4me3) (33). TRExBCBL1-RTA PEL cells, expressing a doxycycline (Doxy)-inducible RTA to allow efficient lytic reactivation, were treated with Doxy for 0 h (latency) or 24 h (lytic replication) and then used to generate ChIP-seq libraries (34). ChIP-seq analysis of the H3K27ac enrichment in the intergenic and intronic regions detected over 19,579 distal active enhancer elements in latent TRExBCBL1-RTA cells, whereas a majority of those active enhancer elements were rapidly depleted upon RTA-induced reactivation (Fig. 1A). ChIP-seq analysis of the H3K4me1 enrichment exhibited a similar pattern (Fig. 1B). Immunoblotting showed that the global levels of histone 3 (H3) expression as well as H3K4me1 and H3K4me3 modification levels were not affected upon RTA-induced reactivation, whereas the H3K27ac modification level was detectably reduced (SI Appendix, Fig. S1A).
Fig. 1.
Identification of active enhancers using H3K27ac and H3K4me1 ChIP-seq. (A, Left) Heatmap of H3K27ac ChIP-seq signals at 19,579 putative active enhancers in TRExBCBL1-RTA cells during latency and lytic replication induced by doxycycline for 24 h; each row represents one enhancer region. (Right) Density plot of average ChIP-seq signals in 10-kb windows around the center of enhancer for latent (green) and lytic (orange) cells. (B) H3K4me1 ChIP-seq signals at 19,579 putative active enhancers as identified in A. (C) DNA binding motifs enriched at superenhancers that were significantly down-regulated upon reactivation. (D) Rank order of H3K27ac ChIP-seq signals. Superenhancers are the population on the Right side of the dotted line (with slope of more than 1). A total of 836 superenhancers are identified and annotated to their nearest genes. Proto-oncogenes and B cell transcription factors important for PEL cells are indicated.
HOMER motif sequence analysis of the enhancer elements whose activities were drastically depleted upon KSHV reactivation predicted involvement of several key B cell transcription factors (Fig. 1C) such as IFN regulatory factor 2/4 (IRF2/4), and YY1 and CTCF chromatin looping factors. Based on the dense occupancy of H3K27ac modification, HOMER analysis also predicted 836 potential superenhancer elements from latent TRExBCBL1-RTA cells. Interestingly, high-ranked superenhancer elements were primarily linked to genes that are essential for B cell development or oncogenesis including MYC, IRF1/4, XBP1, and CD70 (Fig. 1D) (SI Appendix, Fig. S1B). These results collectively indicate that KSHV reactivation leads to the global reduction of host enhancer elements as well as the specific reduction of superenhancer elements of B cell transcription factors and oncogenes.
Identification of Nascent Viral and Host Gene Transcription by GRO-Seq Analysis.
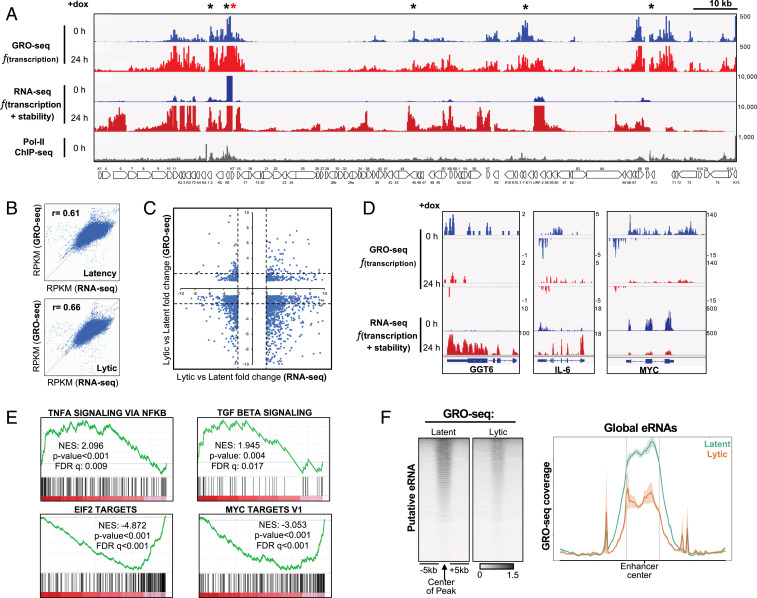
GRO-seq is a derivative of RNA sequencing (RNA-seq) that aims to measure rates of transcription instead of steady-state RNA levels by directly measuring nascent RNA production (35). Specifically, GRO-seq maps the binding sites of actively transcribing RNA polymerase II (RNAPII) at both promoters and enhancers, allowing the capture of unstable and lowly expressed transcripts, such as enhancer RNAs (5). To identify nascent mRNAs produced during latency or lytic replication, mock- or Doxy-treated TRExBCBL1-RTA cells were subjected to GRO-seq analysis. To investigate the correlation between nascent transcripts and steady-state transcripts, we reanalyzed and compared the previously published mRNA-seq data of TRExBCBL1-RTA cells with our GRO-seq results (25). When aligned to the KSHV genome, distinct patterns of viral transcripts between GRO-seq and RNA-seq were observed: GRO-seq detected considerably more viral transcripts during latency than RNA-seq (Fig. 2A), and those transcript peaks were highly correlated to the RNAPII binding sites. This confirms that GRO-seq measures active transcription from the loci where RNAPII is already bound (Fig. 2A, marked in black asterisks). GRO-seq showed similar KSHV transcription patterns between latency and lytic replication, but the only differences were primarily higher transcription profiles during lytic replication than during latency (Fig. 2A). On the other hand, RNA-seq showed considerable difference in KSHV transcription patterns between latency and lytic replication: low expressions of the latent genes except PAN RNA (marked in red asterisk) during latency and high induction of all viral genes during lytic replication (Fig. 2A).
Fig. 2.
Identification of nascent host and viral RNA from GRO-seq. (A) GRO-seq and RNA-seq signals aligned to the KSHV genome during latency and upon reactivation. RNAPII ChIP-seq enrichments are from latency. Black asterisks indicate the regions where the GRO-seq signals are significantly different from RNA-seq signals, and red asterisk is PAN RNA locus that shows high up-regulation in both RNA-seq and GRO-seq during latency. (B) Correlation between GRO-seq and RNA-seq during latency (Top) and lytic replication (Bottom) using Pearson’s correlation. (C) Differential gene expression fold change from latency to lytic replication. Correlation plot of RNA-seq vs. GRO-seq. Genes with P value <0.05 and fold change threshold of ±2 from RNA-seq are displayed. Genes with fold change between −2 and 2 are considered no change. The x axis is differential fold change from latency to lytic replication in RNA-seq; y axis is differential fold change in GRO-seq. (D) GRO-seq and RNA-seq results for three representative host genes: GGT6, IL6, and MYC. (E) GSEA of GRO-seq data upon KSHV reactivation. Enrichment scores are indicated in each plot along with P value and false discovery rate (FDR) q-value. Positive (Top two) and negative (Bottom two) NES (normalized enrichment score) values represent gene sets overrepresented in the topmost up- or down-regulated genes, respectively. (F, Left) Heatmap of GRO-seq signals at putative active enhancers previously identified (Fig. 1A). (Right) Density plot of average GRO-seq signals in 10-kb windows around the center of the enhancer.
When the host transcriptome was examined, the correlation coefficients between GRO-seq and RNA-seq were 0.61 and 0.66 in KSHV latent conditions and lytic replication conditions, respectively, suggesting that there are significant differences in levels between newly synthesized transcripts versus steady-state transcripts (Fig. 2B). Then, we surveyed the fold change of each gene in both RNA abundance (RNA-seq) and transcription (GRO-seq) upon reactivation (Fig. 2C). We first identified host coding genes that were significantly regulated in RNA-seq and then checked whether the trend was similar in GRO-seq. RNA-seq analysis showed that similar numbers of host genes were up-regulated (fold change greater than or equal to 2) or down-regulated (fold change less than or equal to −2) for their expressions. In contrast, GRO-seq showed that expressions of a few host genes were up-regulated upon reactivation, whereas expression of most host genes were highly down-regulated. Interestingly, the coding gene expression profile as indicated by the enrichment heatmap at the promoter of GRO-seq upon reactivation was very similar to that of H3K27ac ChIP-seq (SI Appendix, Fig. S2 A and B), indicating the strong relationship between transcription rate and histone acetylation. For instance, GGT6 and IL6 were highly up-regulated upon reactivation from RNA-seq analysis, but GRO-seq analysis showed no increase of their transcription rate (Fig. 2D). As the IL6 gene has been shown to escape KSHV ORF37 RNase-mediated host mRNA shutoff (36), the increase of IL6 mRNA from RNA-seq analysis may not be due to increased transcription, but due to increased mRNA stability. On the other hand, RNA-seq and GRO-seq showed that RTA-induced reactivation led to the drastic reduction of MYC expression at both transcription and mRNA stability level. These results show that RNA-seq results are more due to changes in RNA stability than to changes in transcription, indicating that transcription and transcript turnover are two distinct mechanisms that govern host gene expression changes upon KSHV reactivation.
Furthermore, we utilized gene set enrichment analysis (GSEA) of the GRO-seq results to evaluate the biological relevance of the host gene alteration at the transcriptional level. We first ranked all host genes according to the extent of their fold changes and P value, followed by computing the normalized enrichment scores of each biological pathway. This analysis revealed that upon KSHV reactivation, inflammation pathways, including TNF-α and TFB-β signaling pathways, were significantly up-regulated, while EIF2 and MYC target pathways were markedly down-regulated (Fig. 2E). These data suggest that GRO-seq analysis is able to identify gene networks whose transcriptional rates are specifically regulated upon KSHV reactivation.
Reduction of Global eRNA Expression upon KSHV Reactivation.
eRNAs represent a class of noncoding unstable RNA molecules transcribed from the enhancer regions and are hallmarks of active enhancers as they have been suggested to play a role in transcriptional regulation in cis and trans. Due to their unstable nature, eRNA transcripts are often unable to be captured by traditional RNA-seq. We plotted the GRO-seq densities for human active enhancers that were previously identified from H3K27ac ChIP-seq (Fig. 2F). Statistical analysis showed that ∼50% of active enhancers produced eRNA above background noise. The global level of eRNAs was also significantly reduced upon RTA-induced reactivation, (Fig. 2F), which was correlated with the reduction of H3K27ac and H3K4me1 at active enhancer elements (Fig. 1 A and B). This further indicates that KSHV reactivation results in the global repression of host enhancer activity.
Identification of Potential eRNAs Downstream of MYC TSS in KSHV-Infected PELs.
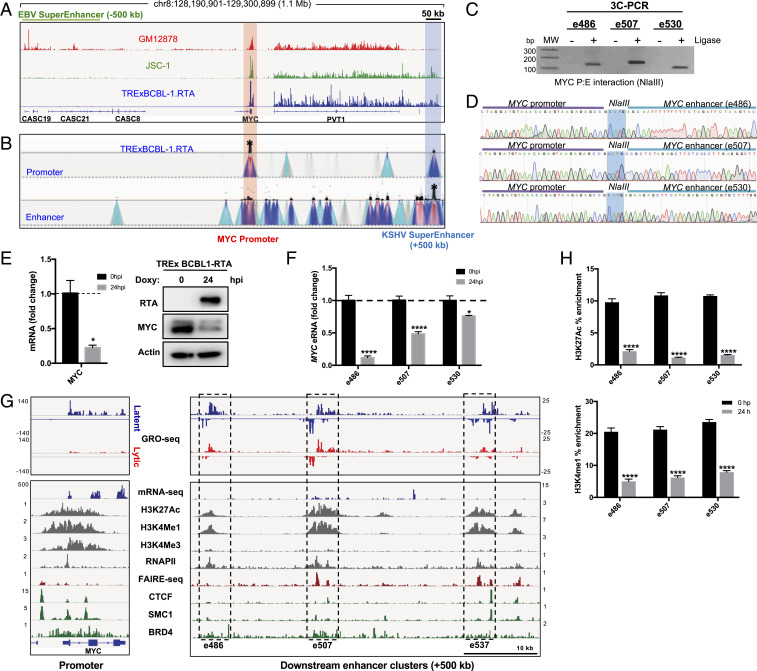
MYC oncogene overexpression is found in many human cancers and often achieved by translocating to another gene’s superenhancers or overactivating its own superenhancers. Specifically, the −500-kb upstream region of the MYC transcription start site (TSS) in EBV + LCLs (GM12878) has been well characterized to function as its superenhancers that are occupied by several EBV latent proteins and specific transcription factors (31). Surprisingly, our GRO-seq analysis showed that unlike EBV + GM12878 LCLs, EBV+/KSHV + JSC-1, and KSHV + TRExBCBL1-RTA PELs both produced strong eRNA-like transcripts in +500 kb downstream of the MYC TSS, downstream of the PVT-1 lncRNA (Fig. 3A). We identified the three highest peaks within the span of 50 kb of the newly identified MYC eRNAs, +486 kb, +507 kb, and +530 kb downstream of the MYC TSS, which were labeled as e486, e507, and e530, respectively (Fig. 3G). To measure the copy number of eRNAs, we performed a drop digital PCR (ddPCR) assay targeting each eRNA and the abundant viral PAN RNA during latency. This showed that the level of GRO-seq peaks appeared to be similar to the copy numbers of three eRNAs: e486 (6.1 copy/ng), e507 (14.8 copy/ng), and e530 (8.4 copy/ng) along with PAN RNA (172,375 copy/ng). This indicates that eRNAs are expressed downstream of the MYC in KSHV-infected PELs, whereas they are expressed upstream of the MYC in EBV+ LCLs.
Fig. 3.
KSHV + PEL cell lines show potential MYC superenhancers downstream of the MYC TSS, which reduces upon KSHV reactivation. (A) GRO-seq peaks near the MYC locus. EBNA2-mediated EBV superenhancer is indicated by high peaks −500 kb upstream of the MYC TSS (red highlight) and potential the KSHV superenhancer is indicated +500 kb downstream (blue highlight). (B) The 4C-seq signals in latent TRExBCBL1-RTA cells near the MYC locus. (Top) MYC promoter set as the 4C viewpoint/anchor sequence (red highlight); (Bottom) enhancer set as the 4C viewpoint/anchor sequence (blue highlight). (C) A 3C-PCR showing intensities of a specific promoter:enhancer (P:E) looping in the MYC locus. Control 3C samples without T4 ligase are shown as “−.” (D) Sanger sequencing of the 3C-PCR products shows that the ligated fragment comprises the regions from MYC promoter and enhancer. (E) qRT-PCR and immunoblot showing reduction of MYC mRNA and protein level upon doxycycline-induced KSHV reactivation. (F) qRT-PCR measuring reduction of enhancer transcripts at each MYC enhancer. (G) Various sequencing results (in-house and public data) confirming the activity of the MYC superenhancer. Each enhancer cluster (black dotted box) is labeled as the distance away from the MYC TSS (+486 kb, +507 kb, +537 kb). GRO-seq peaks are for both latency (blue) and lytic replication (red), but all others show peaks during latency only. (H) H3K27ac and H3K4me1 ChIP-qPCR showing reduction of enhancer activity at MYC superenhancers.
To further test whether these eRNA-expressing enhancer regions function specifically as MYC enhancers, we performed chromosome conformation capture (3C) combined with high-throughput sequencing (4C-seq) on TRExBCBL1-RTA cells to detect the genomic interaction of these two regions. We used either the MYC promoter (forward) or enhancer (backward) as the bait sequence and identified the genomic loci that physically interacted with the bait (Materials and Methods). Both forward and backward 4C-seqs showed that TRExBCBL1-RTA PELs carried specific interactions between the MYC promoter and the downstream enhancers corresponding to the eRNA transcription sites (Fig. 3B). The 3C-PCR and DNA sequence analysis further validated the specific interaction between the MYC promoter and each of its enhancer clusters identified by 4C-seq analysis (Fig. 3 C and D). An in-depth examination of GRO-seq, H3K27ac ChIP-seq, and H3K4me1 ChIP-seq showed extensive overlap at each enhancer cluster (Fig. 3G). By reanalyzing publicly available ChIP-seq data, we observed the colocalizations of RNAPII, CTCF, SMC1, and BRD4 clusters with these enhancer elements, and the formaldehyde-assisted isolation of regulatory elements sequencing (FAIRE-seq) also confirmed chromatin accessibility at these regions (37–40). Active enhancers are marked by bidirectional GRO-seq signal, high H3K27ac and H3K4me1 relative to H3K4me3. These regions need to be accessible as indicated by FAIRE-seq so that RNAPII and other chromatin modulating factors (CTCF, SMC1, and BRD4) can be recruited to the sites. These results strongly indicate that the eRNA-expressing regions downstream of the MYC gene function as its superenhancers. This also suggests that unlike EBV-infected LCL cells, KSHV-infected PEL cells utilize superenhancers downstream of MYC.
Transcriptional Regulation of MYC eRNA Expression upon KSHV Lytic Induction.
The elevated MYC expression has been shown to be necessary to repress KSHV lytic gene expression in PELs, resulting in the maintenance of viral latency (28). Also, RNAi-mediated MYC knockdown induced KSHV reactivation and also reactivation-independent apoptosis, indicating its role in maintaining latency (28). As seen with many host genes, MYC expression was also repressed upon KSHV reactivation at both mRNA and protein levels in TRExBCBL1-RTA PELs (Fig. 3E). qRT-PCR showed that RTA-mediated reactivation led to the pronounced reduction of e486 and e507 eRNA transcripts and modest reduction of e530 eRNA transcript (Fig. 3F). GRO-seq also showed visible reductions of all three eRNA peaks (Fig. 3G). ChIP-qPCR also showed that RTA-mediated reactivation led to the marked reduction of H3K27ac and H3K4me1 enhancer mark occupancy at the MYC superenhancer regions (Fig. 3H). These results collectively demonstrate that KSHV reactivation represses MYC expression to facilitate lytic gene activation, and its expression may partially be controlled by the reduced superenhancer activity.
Disruption of MYC Superenhancers Leads to MYC Reduction and KSHV Reactivation.
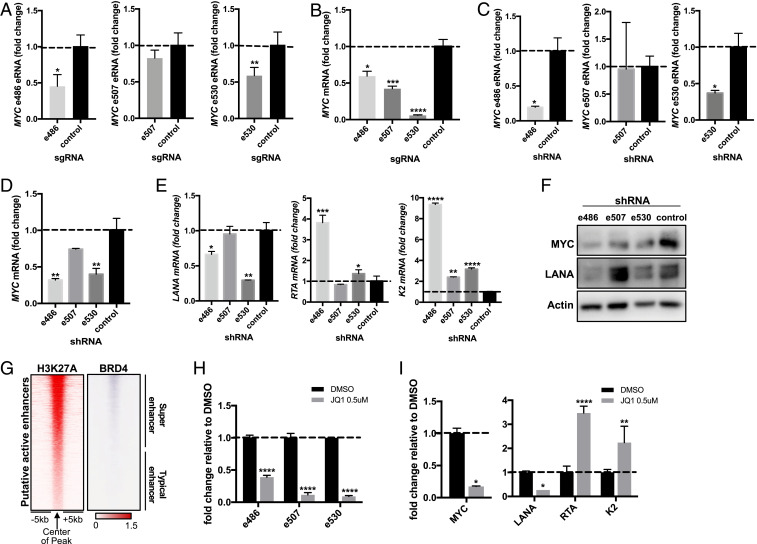
To assess the functionality of the MYC enhancers, we used the dCas9-KRAB CRISPR inhibition (CRISPRi) approach. TRExBCBL1-RTA cells were transduced with lentivirus expressing the nuclease-inactive dCas9-KRAB fusion repressor together with sgRNAs targeting each MYC enhancer (41). We also used sgRNAs targeting MYC mRNA and PVT-1 ncRNA as controls to ensure the efficacy of this approach (SI Appendix, Fig. S3). e486- or e530-specific CRISPRi reduced e486 or e530 expression level, respectively, while e507-specific CRISPRi showed minimal effect on e507 level (Fig. 4A). qRT-PCR showed that the CRISPRi-mediated inhibition of the eRNA transcription significantly reduced MYC mRNA expression compared to the control CRISPRi treatment (Fig. 4B).
Fig. 4.
Disruption of MYC superenhancers leads to KSHV reactivation. (A) dCas9-KRAB-sgRNA-mediated CRISPRi targeting each MYC eRNA, and dCas9-KRAB alone used as control. (B) qRT-PCR of host MYC mRNA upon CRISPR inhibition of the three MYC eRNAs. (C) shRNA-mediated RNA interference (RNAi) targeting each MYC eRNA, and scrambled sequence used as control. (D and E) qRT-PCR of MYC and viral gene mRNAs (LANA, RTA, and K2) upon shRNA transduction. (F) Immunoblot for MYC and LANA upon shRNA transduction validates the qRT-PCR results. (G) BRD4 ChIP-seq signals from the host genome during latency are aligned next to H3K27ac signals. BRD4 preferentially binds to superenhancers but not typical enhancers. (H) qRT-PCR results for MYC eRNA upon treatment with 0.5 μM of JQ1 for 24 h. (I) qRT-PCR results for MYC mRNA and viral mRNAs (LANA, RTA, and K2).
In order to further confirm the functional role of MYC eRNAs, TRExBCBL1-RTA PELs were transduced with lentiviruses expressing scrambled control shRNA or eRNA-specific shRNA, selected for puromycin resistance to remove untransduced cells, and then harvested to extract total RNAs. e486- or e530-specific shRNA efficiently knocked down the corresponding eRNA, whereas e507-specific shRNA showed minimal knockdown of e507 (Fig. 4C). qRT-PCR showed that depletion of either e486 or e530 eRNA expression significantly reduced MYC mRNA expression at different magnitudes (Fig. 4D). Similar to the e507-specific CRISPRi, the e507-specific shRNA weakly reduced e507 level but both treatments led to the strong reduction of MYC mRNA (Fig. 4 B, D, and F). Subsequently, the e486- or e530-specific shRNA led to the reduction of LANA latent gene expression and induction of RTA and K2 lytic gene expression (Fig. 4E), indicating that knockdown of these eRNAs prompts KSHV lytic gene expression. Immunoblotting also showed the reduction of MYC and LANA levels (Fig. 4F). These studies collectively indicate that depletion of the enhancer function and eRNA expression lead to repression of the MYC expression that ultimately affects KSHV gene regulation.
BRD4 (bromodomain-containing protein 4) is a member of the BET (bromodomain and extra terminal domain) family and an important coactivator that binds to H3K27ac specifically on superenhancers but not on typical enhancers and recruits RNAPII to trigger transcription (Fig. 4G) (42, 43). JQ1 is a well-known small molecule inhibitor that prevents BRD4 from associating with H3K27ac at superenhancers to suppress transcription (44). To test the effects of JQ1 on the MYC eRNA transcription, TRExBCBL1-RTA PELs were treated with 0.5 μM JQ1 or dimethyl sulfoxide (DMSO) control for 24 h and the expressions of the MYC eRNAs were assessed by qRT-PCR. Consequently, JQ1 treatment markedly repressed all three MYC eRNA expressions, resulting in the reduction of MYC expression and the induction of KSHV lytic gene expression (Fig. 4 H and I). These results indicate that JQ1-mediated BRD4 perturbation disrupts the MYC superenhancer function, reducing both MYC eRNA and mRNA levels while inducing KSHV lytic gene expressions.
Potential Roles of Cellular IRF4 and Viral IRF4 in MYC eRNA Expression.
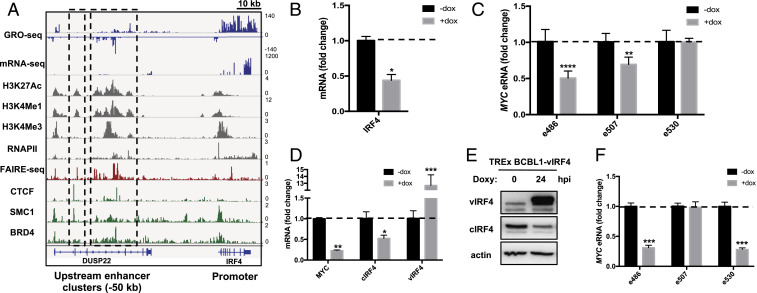
Host IFN regulatory factor (IRF4) is another essential gene that was identified by H3K27ac ChIP-seq to be regulated by superenhancers (Fig. 1D). Its enhancer clusters were found in the intragenic region of DUSP22 gene upstream (Fig. 5A). We further investigated the role of IRF4 in the enhancer regulation of PEL cells because not only is IRF4 essential for PEL growth and survival (45, 46), IRF4 itself also directly binds to enhancers (47). Because IRF4 was one of the top enriched factors in superenhancers as identified by the HOMER motif sequence analysis (Fig. 1C), and because it has been known to directly target MYC expression, we examined the potential role of IRF4 in MYC eRNA expression. BCBL-1 PELs carrying Doxy-inducible IRF4-specific shRNA were treated with a low concentration of Doxy (100 ng/mL) for 72 h and then tested for cellular IRF4 expression and MYC eRNA expression by qRT-PCR (46). This showed that IRF4-specific shRNA treatment detectably reduced IRF4 expression (Fig. 5B) as well as MYC e486 and e507 eRNA expressions (Fig. 5C), suggesting the role of IRF4 in MYC eRNA expression.
Fig. 5.
Role of IRF4 in PEL enhancer activity. (A) Identification of IRF4 superenhancer −50 kb upstream of the IRF4 TSS by aligning multiple different sequencing results. The two enhancer clusters (black dotted boxes) are embedded within the intronic region of DUSP22. (B) qRT-PCR of IRF4 in BCBL-1 cells with doxycycline-inducible shRNA targeting IRF4. (C) qRT-PCR of MYC eRNA. Reduction of IRF4 expression leads to down-regulation of e486 and e507 but not e530. (D) qRT-PCR of MYC, cellular IRF4, and viral IRF4 mRNA in TRExBCBL1-vIRF4 treated with doxycycline for 24 h to induce viral IRF4 expression. (E) Immunoblot showing induction of vIRF4 and reduction of cellular IRF4. (F) qRT-PCR results showing the reduction of eRNAs upon vIRF4 overexpression.
We have previously shown that the KSHV viral IRF4 (vIRF4) lytic protein robustly suppresses expression of host or cellular IRF4 (cIRF4) at the transcriptional level to reshape host gene expression profiles and facilitate viral lytic replication (48). TRExBCBL1-vIRF4 PELs carrying Doxy-inducible vIRF4 expression were treated with Doxy for 24 h and subjected to qRT-PCR analysis. vIRF4 overexpression alone robustly reduced cIRF4 and MYC expression as expected (Fig. 5 D and E). Also, a significant repression of MYC e486 and e530 eRNAs was observed, indicating that the vIRF4-mediated MYC reduction may be regulated at the superenhancer level. Collectively, these results suggest that during KSHV latency, cellular IRF4 regulates MYC superenhancers to establish viral latency and to support oncogenesis. Upon viral reactivation, the expression of lytic viral IRF4 represses cellular IRF4 expression, thereby dysregulating the superenhancer-mediated MYC expression.
Discussion
This study introduces a layer of epigenomic regulation in KSHV-infected PEL cells and suggests a potential mechanism of how KSHV reactivation changes the host epigenome. We combined ChIP-seq, GRO-seq, and 4C-seq datasets with publicly available Next Generation Sequencing datasets to generate an in-depth genome-wide analysis of host enhancers and superenhancers during latent infection and lytic replication of KSHV. Firstly, we identified that a number of active enhancer elements were detected during KSHV latency and those enhancer elements were predicted to be targeted by IRF2 and IRF4 B cell transcription factors and YY1 and CTCF chromatin looping factors. Upon RTA-mediated KSHV lytic replication, however, host enhancer activity was globally reduced as indicated by the decreased level of H3K27ac and H3K4me1 modification. p300 transcriptional cofactor contains histone acetyltransferase (HAT) activity that is responsible for H3K27ac modification (49). We have previously shown that KSHV encodes viral IRF1 lytic protein that directly binds to p300 and displaces p300/CBP-associated factor from p300 complexes (50, 51). This interaction inhibits p300 HAT activity, resulting in the drastic reduction of global H3K27ac and the alteration of host chromatin structure. On the other hand, it has not been studied how KSHV directly or indirectly controls H3K4me1 modification. Nevertheless, this suggests that KSHV has evolved to carry a number of viral proteins to target host epigenetic factors, resulting in the alteration of host enhancer activity for its lifecycle.
Intriguingly, GRO-seq patterns of viral transcription were highly similar between KSHV latency and lytic replication, indicating that the RNAPII occupancy is not significantly changed on the viral genome upon reactivation. In fact, we have previously shown that the KSHV genome is bivalent as both active and repressive histone marks are associated with the viral genome (22). Furthermore, RNAPII is already recruited to the viral loci but its transcription is paused by the host’s negative elongation factor, which is necessary to efficiently respond to environmental stimuli and promptly activate viral lytic gene expression (52).
GRO-seq measures nascent RNA levels, whereas RNA-seq measures steady-state RNA levels. Both GRO-seq and RNA-seq analyses showed similar viral transcription profiles during lytic replication except for a few genes, such as ORF6 and ORF59, which were more highly expressed from RNA-seq than from GRO-seq, indicating high stability of these genes. Similarly, expression of several host genes such as GGT6 and IL6 were highly up-regulated upon KSHV reactivation from RNA-seq analysis, but they showed little or no increase from GRO-seq analysis. Previous studies have shown that lytic protein ORF37 is primarily responsible for host mRNA shutoff upon reactivation but the host IL6 gene can escape ORF37-mediated host mRNA shutoff (53). Thus, the difference between RNA-seq results and GRO-seq results suggests that transcription and transcript turnover are two potential mechanisms that govern host and viral gene expressions upon KSHV reactivation.
We identified KSHV-specific superenhancer elements and corresponding eRNAs of MYC. While the MYC superenhancer elements were found upstream of its TSS in EBV + LCLs, they were found downstream in KSHV + infected or EBV+/KSHV + coinfected PELs. This may be due to the lack of certain EBV EBNAs that derive EBV-specific superenhancer (31), which suggests that in PELs, MYC enhancer regulation is determined by KSHV factors.
KSHV reactivation led to down-regulation of the MYC eRNA expression and enhancer activity, which ultimately lowered MYC gene expression. Similarly, JQ1-mediated BRD4 perturbation disrupted the MYC superenhancer function, reducing both MYC eRNA and mRNA levels. It should be noted that while RNAi- or CRISPRi-mediated repression of eRNA expression reduced MYC expression and altered KSHV gene expression, the effects were highly variable potentially due to the transduction efficiency.
Cellular IRF4 was identified to be one of the most essential genes in PEL cell growth and survival (45). Our results show that not only is cIRF4 an essential oncogene that is regulated by superenhancers, its protein also binds to many other superenhancers that are most significantly dysregulated by reactivation, thus confirming its crucial role in PEL. Both shRNA-mediated and vIRF4-mediated cIRF4 suppression resulted in the reduction of MYC eRNAs, although at different levels. This suggests that each eRNA expression may depend on the level of involvement and binding affinity of several transcription factors.
Since host reprogramming is intricately regulated by viruses, it is possible that several different viral factors and pathways are involved in host enhancer modulation. Another mechanism could be a contribution from the highly abundant viral PAN RNA which interacts with several chromatin modification complexes to regulate gene expression. As several nuclear lncRNAs have been found to act as eRNA and target enhancers in trans (54, 55), it is possible that the multifunctional viral ncRNA may directly or indirectly modulate eRNA expression. A recent study has shown that some eRNAs are not limited to the nucleus and may function in the cytoplasm (56). In this case, these eRNAs may also be targeted by KSHV ORF37 RNase. Taken together, there are multiple ways that virus may control eRNA expression to regulate superenhancer activity. Nevertheless, our analyses identify eRNAs and related transcription factors that are highly correlated with KSHV gene expression, thus highlighting the importance of transcriptional and epigenomic dynamics for KSHV lifecycle. Furthermore, our study suggests a mechanism of how KSHV modulates host enhancer repertoire at different stages of the lifecycle to benefit its own replication and immune modulation.
Materials and Methods
Cell Culture.
TRExBCBL1 cell lines contain a doxycycline-inducible gene that can be induced with 1 μg/mL Doxy to overexpress RTA or vIRF4. All Doxy treatments lasted for 24 h. They were cultured in RPMI 1640 media supplemented with 10% Tet system-approved fetal bovine serum (FBS) (Clontech), 100 U/mL penicillin, 100 μg/mL streptomycin, and 20 μg/mL hygromycin B. JSC-1 was grown in RPMI 1640 media supplemented with 10% FBS (Clontech), 100 U/mL penicillin, and 100 μg/mL streptomycin. BCBL-1 with Doxy-inducible shIRF4 or sh-scrambled control (gift from Saumendra Sarkar, University of Pittsburgh, Pittsburgh, PA) were grown in RPMI 1640 media supplemented with 10% FBS (Clontech), 100 U/mL penicillin, and 100 μg/mL streptomycin, and selected with 1 μg/mL puromycin.
Reagents and Antibodies.
The following antibodies were used for immunoblot or for ChIP: rabbit anti-histone H3 (Abcam 1791), rabbit anti-H3K27ac (Active Motif 39133), rabbit anti-H3K4me1 (Abcam 8895), rabbit anti-H3K4me3 (Millipore 04-745), rabbit anti-MYC (Abcam 32072), mouse anti-β-actin (Santa Cruz 47778), rat anti-LANA (Advanced Biotech 13-210-100), rabbit vIRF4 serum (Covance). Rabbit anti-RTA was a gift from Yoshihiro Izumiya, University of California, Davis, CA).
qRT-PCR.
Total RNA was extracted using Tri reagent (Sigma). A total of 1 μg of RNA was used for DNase I (Sigma), reverse transcribed with iScript cDNA synthesis kit (Bio-Rad), and the resulting cDNA was used for qPCR. SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) was used for qPCR according to the manufacturer’s instructions. Each sample was normalized to 18S, and the ddCt fold change method was used to calculate relative quantification. Primer sequences used for qRT-PCR are found in SI Appendix, Table S1.
ChIP-qPCR.
Roughly 10 μg of chromatin was used for each ChIP. TRExBCBL1-RTA cells were treated with 1 μg/mL Doxy for 0 or 24 h, and cross-linked with 1% formaldehyde for 10 min at room temperature (RT) then quenched using 0.125 M glycine. Cells were washed with ice-cold 1× PBS then resuspended in ice-cold swelling buffer (5 mM Pipes pH 8.0, 85 mM KCl, 1% Igepal). Cells were then homogenized using a glass douncer, then incubated in nuclei lysis buffer (50 mM Tris-Cl pH 8.1, 10 mM ethylenediaminetetraacetic acid [EDTA], 1% sodium dodecyl sulfate [SDS]) plus protease inhibitor on ice for 30 min, then subjected to sonication using Bioruptor Pico (Diagenode). The nuclei lysates were then diluted at least fivefold with ice-cold IP buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Igepal, 0.25% deoxycholic acid, 1 mM EDTA) with protease inhibitor, then incubated with antibody of interest overnight in 4 °C. Magnetic protein A/G beads (Pierce) were used to pull down each ChIP for 2 h at 4 °C, then the immunoprecipitant was washed twice with IP wash buffer 1 (same as IP dilution buffer, without the protease inhibitor) then twice with IP wash buffer 2 (100 mM Tris-Cl pH 9.0, 500 mM LiCl, 1% Igepal, 1% deoxycholic acid), both at RT. The final antibody/chromatin complexes were eluted using IP elution buffer. After elution, DNA was purified using MinElute PCR clean up kit (Qiagen) and subjected to qPCR or further processed as ChIP-seq library. Primer sequences used for ChIP-qPCR are found in SI Appendix, Table S1.
ChIP-Seq.
All ChIP-seq libraries were prepared using KAPA Hyper Prep Kit (KAPA KK8503) according to the manufacturer’s instructions. Samples were sequenced on a Illumina NextSeq550 machine using 75-bp single-ended reads.
GRO-Seq.
GRO-seq experiments were performed as previously reported (57). Briefly, 40 million TRExBCBL1-RTA cells during latency and upon reactivation were washed with ice-cold PBS 3 times, incubated with swelling buffer (10 mM Tris-Cl pH7.5, 2 mM MgCl2, 3 mM CaCl2) for 5 min on ice, harvested, and lysed in lysis buffer (swelling buffer plus 0.5% Nonidet P-40 and 10% glycerol). The resultant nuclei were washed one more time with 10 mL lysis buffer and finally resuspended in 100 μL of freezing buffer (50 mM Tris-Cl pH8.3, 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA). For the run-on assay, resuspended nuclei were mixed with an equal volume of reaction buffer (10 mM Tris-Cl pH 8.0, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 300 mM KCl, 20 units of SUPERase-IN, 1% sarkosyl, 500 μM adenosine-5'-triphosphate [ATP], guanosine-5'-triphosphate [GTP], and 5-bromouridine-5′-triphosphate [Br-UTP], 2 μM cytidine 5′-triphosphate [CTP]) and incubated for 5 min at 30 °C. The resultant nuclear run-on RNA (NRO-RNA) was then extracted with TRIzol LS reagent (Life Technologies) following manufacturer’s instructions. NRO-RNA was fragmented to ∼300 to 500 nt by alkaline base hydrolysis on ice and followed by treatment with DNase I and Antarctic phosphatase. These fragmented Br-UTP-labeled nascent RNA were then immunoprecipitated with anti-BrdU agarose beads (SC32323ac, Santa Cruz Biotechnology) in binding buffer (0.5× saline-sodium phosphate-EDTA [SSPE], 1 mM EDTA, 0.05% Tween) for 3 h at 4 °C with rotation. Subsequently, T4 polynucleotide kinase (T4 PNK) was used to repair the end of the immuneoprecipitated BrU-NRO-RNA, at 37 °C for 1 h. The RNA was extracted and precipitated using acidic phenol-chloroform. cDNA synthesis was performed as per a published method (58) with few modifications. The RNA fragments were subjected to poly-A tailing reaction by poly-A polymerase (NEB) for 30 min at 37 °C. Subsequently, reverse transcription was performed using oNTI223 primer (5′-/5Phos/GA TCG TCG GAC TGT AGA ACT CT/idSp/CAA GCA GAA GAC GGC ATA CGA TTT TTT TTT TTT TTT TTT TTV N-3′) and superscript III RT kit (Life Technologies). The cDNA products were separated on a 10% polyacrylamide tris-borate-EDTA (TBE)-urea gel and only those migrated between ∼100 to 500 bp were excised and recovered by gel extraction. After that, the first-strand cDNA was circularized by CircLigase (Epicentre) and relinearized by APE1 (NEB). Relinearized single-strand cDNA (sscDNA) was separated by a 10% polyacrylamide TBE gel as described above and the product of needed size was excised (∼170 to 400 bp) for gel extraction. Finally, sscDNA template was amplified by PCR (usually between 10 and 14 PCR cycles) using the Phusion High-Fidelity enzyme (NEB) according to the manufacturer’s instructions. Final libraries were sequenced on the Illumina Hi-SEq. 2500 using single-read 100-cycle runs.
ddPCR.
ddPCR was carried out using the Bio-Rad system. First, total RNAs were reverse transcribed using DNase I (Sigma), then reverse transcribed with iScript cDNA synthesis kit (Bio-Rad). Droplet formation was carried out using ddPCR Supermix for Probes (Bio-Rad, No. 1863023) and QX200 droplet generator (Bio-Rad) according to the manufacturer’s instructions. The generated oil droplets were transferred onto a 96-well plate, and subsequently PCR was carried out using a QX200 reader (Bio-Rad). Data from the droplet reader were given as copies per nanogram of RNA for accurate quantification. Totals of 40 ng and 40 pg were used for one ddPCR reaction for eRNA and PAN RNA, respectively. All primers and probes were synthesized by Integrated DNA Technologies (IDT). Hydrolysis probes contained a 5′-FAM fluorophore and a 3′-Iowa black quencher. Primer sequences are the same as qRT-PCR primers, and probes for the genes studied are indicated below (5′ to 3′):
e486_Probe: AGCCACAGTGTTCTGGATGTCCTT
e507_Probe: ACGTGACTCCAATTCATCCCACAGT
e530_Probe: AGAAGTCACAGTTACACAAGAGGTACA
PANRNA_Probe: CATTGGACTAAAGTGGTGTGCGGC.
4C-Seq.
4C-seq experiments largely followed a published protocol (59) with modification. Briefly, 10 million cells were cross-linked with 1% formaldehyde for 10 min and nuclei were extracted. Nuclei were resuspended in restriction enzyme buffer and incubated with 0.3% SDS for 1 h at 37 °C and further incubated with 2% Triton X-100 for 1 h. A total of 400 U of NlaIII restriction enzyme was added and incubated overnight. Restriction enzyme was heat inactivated at 65 °C for 20 min. Ligation of DNA regions in close physical proximity was performed using 1,000 U of T4 DNA ligase (NEB) overnight. After de-cross-linking, the second digestion and ligation were performed using restriction enzyme DpnII and T4 DNA ligase. 4C-seq libraries were amplified using first round of PCR with the primers indicated in SI Appendix, Table S2. Primers contained Illumina sequencing adaptors, and second round of PCR primers contained Index sequences (NEBNext Multiplex Oligos for Illumina; New England Biolabs Inc). Samples were sequenced on an Illumina NextSeq550 machine using 75-bp single-ended reads.
3C-PCR.
3C-PCR followed previous methods (60). In short, it was performed using NlaIII restriction enzyme, using primers tested for their efficiency and linearity (primers listed in SI Appendix, Table S2. For results presented, 35 cycles of PCR were performed.
RNA-Seq Analysis.
TRExBCBL1-RTA RNA-seq data (0 h and 24 h) were downloaded from National Center for Biotechnology Information (NCBI) (GSE123898) and aligned to either human hg19 or KSHV JSC-1 BAC16 (GQ994935.1) reference genomes. Aligned.bam files were visualized using an Integrative Genomics Viewer (IGV). Correlation scatterplots comparing RNA-seq and GRO-seq were done using R and the ggplots2 package.
ChIP-Seq Analysis.
H3K27ac and H3K4me1 ChIP-seq were produced in-house, while the rest were downloaded from NCBI (H3K4me3, GSM1265857; BRD4, GSM2769881; RNAPII, GSM1265864; CTCF, GSM941710; and SMC1, GSM941711). The raw files were initially trimmed and aligned to the hg19 genome using Partek Flow and bowtie2. Reads were trimmed based on quality score from both the 5′ and 3′ ends. FAIRE-seq data were analyzed similarly (GSM1223899). The resulting aligned.bam files were either visualized with the IGV or used for further analysis using HOMER (Hypergeometric Optimization of Motif EnRichment). Tag directories were created from the.bam files using the standard settings. Peak calling for transcription factors, histone markers, and superenhancers utilized the “factor,” “histone,” and “super” style options, respectively, with cumulative Poisson P value requirement of 0.0001 P value for removing tag counts that are not statistically significant. For H3K27ac and H3K4me1 modification, the peak size setting was set to “-size 1000.” The peaks were annotated using the hg19 genome for HOMER and the resulting.txt file was imported into Excel. For H3K27ac ChIP-seq, potential active enhancers were found by removing any peaks annotated to the transcription start site. The resulting differential peak files were then used for motif analysis in HOMER or to generate heatmaps and profiles using NGSPlot.
GRO-Seq Analysis.
Initial trimming and aligning were done in the same manner as ChIP-seq data. Tag Directories were created from the.bam files with standard settings and peaks were identified using the “-style groseq” option. Peaks were annotated to remove the transcription start site-associated peaks and used for NGSPlot to generate heatmaps and profiles.
4C-Seq Analysis.
For all 4C-seq data, the raw files were directly used for analysis with 4cseqpipe. The hg19 genome was used with the linear mean stat type and trend resolution of 1,000.
CRISPR Interference.
Two small guide RNAs (sgRNAs) were designed for each targeted locus using the Alt-R Custom Cas9 crRNA Design Tool (IDT) and cloned into the lentiviral vector pLKO.1-U6-2sgRNA-ccdB-EF1a-puromycin (a gift for Xingxu Huang, Shanghaitech University, Shanghai, China) with the strategy developed by Xingxu Huang’s laboratory. Lentiviral gRNAs or Lenti-dCas9-KRAB-blast plasmids (89567, Addgene) were cotransfected with packaging plasmids into HEK293T cells using polyethylenimine (PEI). Culture medium containing lentivirus particles for gRNA and dCas9-KRAB was harvested after 2 d, and TRExBCBL1-RTA cells were spin-infected with lentiviruses containing 10 µg/mL polybrene (Sigma) and no antibiotics at 450 × g for 90 min at 32 °C. Fresh media were added to the cells and the cells were incubated in 37 °C for 2 d before being selected with 1 μg/mL puromycin to select sgRNA-expressing cells. Cells were then harvested 2 d after selection. sgRNA sequences are included in SI Appendix, Table S3.
shRNA Lentivirus Transduction.
Lentiviruses were produced in HEK293T cells using the pLKO shRNA system with packaging plasmids using PEI. TRExBCBL1-RTA cells were spin-infected with lentiviruses containing 10 µg/mL polybrene (Sigma) and no antibiotics at 450 × g for 90 min at 32 °C. Fresh media were added to the cells and they were incubated in 37 °C for 2 d before being selected with 1 μg/mL puromycin. Cells were then harvested 2 d after selection. shRNA sequences are included in SI Appendix, Table S3.
Statistical Analyses.
The statistical tests were calculated using GraphPad Prism v.6.0 (GraphPad Software). All data represent at least two independent experiments. Analyses include one-way ANOVA with Dunnett’s comparison and two-way ANOVA with Bonferroni’s comparison for multicomponent comparisons. Two-tailed unpaired Student’s t test was used for two-component comparisons. Data are presented as the mean ± SD. Asterisks indicate statistically significant differences between groups and across time points as determined by t test. * indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001, and **** indicates P < 0.0001.
Supplementary Material
Acknowledgments
We thank Drs. Yoshihiro Izumiya, Saumendra Sarkar, and Xingxu Huang for providing reagents. This work was partly supported by NIH grants (CA200422, CA251275, CA250052, AI116585, AI140718, AI140705, DE023926, DE027888, DE028521), and the Fletcher Jones Foundation grant (J.U.J.).
Footnotes
Competing interest statement: J.U.J. is a scientific adviser of Vaccine Stabilization Institute, a California corporation.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922216117/-/DCSupplemental.
Data Availability.
The GRO-seq, 4C-seq, and H3K27ac ChIP-seq data were deposited in NCBI Gene Expression Omnibus (GEO) database under accession number GSE147063 (61). Previously reported ChIP-seq data (H3K4me3, BRD4, RNAPII, CTCF, SMC1), FAIRE-seq data, and RNA-seq data were used to support this study and are available in NCBI GEO database under accession numbers GSM1265857, GSM2769881, GSM1265864, GSM941710, GSM941711, and GSE123898, respectively (62–68). These datasets are cited at relevant places within the text.
References
- 1.Heinz S., Romanoski C. E., Benner C., Glass C. K., The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 16, 144–154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Creyghton M. P. et al., Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vähärautio A., Taipale J., Cancer. Cancer by super-enhancer. Science 346, 1291–1292 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Hnisz D. et al., Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lam M. T., Li W., Rosenfeld M. G., Glass C. K., Enhancer RNAs and regulated transcriptional programs. Trends Biochem. Sci. 39, 170–182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu F., Enhancer-derived RNA: A primer. Genomics Proteomics Bioinformatics 15, 196–200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balakrishnan L., Milavetz B., Epigenetic regulation of viral biological processes. Viruses 9, 346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milavetz B. I., Balakrishnan L., Viral epigenetics. Methods Mol. Biol. 1238, 569–596 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman P. M., Epigenetics and genetics of viral latency. Cell Host Microbe 19, 619–628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cesarman E. et al., In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86, 2708–2714 (1995). [PubMed] [Google Scholar]
- 11.Chang Y. et al., Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 266, 1865–1869 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Nador R. G. et al., Primary effusion lymphoma: A distinct clinicopathologic entity associated with the Kaposi’s sarcoma-associated herpes virus. Blood 88, 645–656 (1996). [PubMed] [Google Scholar]
- 13.Cesarman E., Gammaherpesviruses and lymphoproliferative disorders. Annu. Rev. Pathol. 9, 349–372 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Giffin L., Damania B., KSHV: Pathways to tumorigenesis and persistent infection. Adv. Virus Res. 88, 111–159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang J., Renne R., Dittmer D., Ganem D., Inflammatory cytokines and the reactivation of Kaposi’s sarcoma-associated herpesvirus lytic replication. Virology 266, 17–25 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Cavallin L. E., Goldschmidt-Clermont P., Mesri E. A., Molecular and cellular mechanisms of KSHV oncogenesis of Kaposi’s sarcoma associated with HIV/AIDS. PLoS Pathog. 10, e1004154 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aneja K. K., Yuan Y., Reactivation and lytic replication of Kaposi’s sarcoma-associated herpesvirus: An update. Front. Microbiol. 8, 613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu F. et al., Systematic identification of cellular signals reactivating Kaposi sarcoma-associated herpesvirus. PLoS Pathog. 3, e44 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glaunsinger B., Chavez L., Ganem D., The exonuclease and host shutoff functions of the SOX protein of Kaposi’s sarcoma-associated herpesvirus are genetically separable. J. Virol. 79, 7396–7401 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glaunsinger B., Ganem D., Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell 13, 713–723 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Toth Z., Brulois K., Jung J. U., The chromatin landscape of Kaposi’s sarcoma-associated herpesvirus. Viruses 5, 1346–1373 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toth Z. et al., Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 6, e1001013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Günther T., Grundhoff A., The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 6, e1000935 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellert J. et al., A structural basis for BRD2/4-mediated host chromatin interaction and oligomer assembly of Kaposi sarcoma-associated herpesvirus and murine gammaherpesvirus LANA proteins. PLoS Pathog. 9, e1003640 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papp B. et al., Genome-wide identification of direct RTA targets reveals key host factors for Kaposi’s sarcoma-associated herpesvirus lytic reactivation. J. Virol. 93, e01978-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer N., Penn L. Z., Reflecting on 25 years with MYC. Nat. Rev. Cancer 8, 976–990 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Katano H., Sato Y., Sata T., Expression of p53 and human herpesvirus-8 (HHV-8)-encoded latency-associated nuclear antigen with inhibition of apoptosis in HHV-8-associated malignancies. Cancer 92, 3076–3084 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Li X., Chen S., Feng J., Deng H., Sun R., Myc is required for the maintenance of Kaposi’s sarcoma-associated herpesvirus latency. J. Virol. 84, 8945–8948 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baresova P., Pitha P. M., Lubyova B., Kaposi sarcoma-associated herpesvirus vIRF-3 protein binds to F-box of Skp2 protein and acts as a regulator of c-Myc protein function and stability. J. Biol. Chem. 287, 16199–16208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Martin H. J., Liao G., Hayward S. D., The Kaposi’s sarcoma-associated herpesvirus LANA protein stabilizes and activates c-Myc. J. Virol. 81, 10451–10459 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou H. et al., Epstein-Barr virus oncoprotein super-enhancers control B cell growth. Cell Host Microbe 17, 205–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang J. et al., Epstein-Barr virus super-enhancer eRNAs are essential for MYC oncogene expression and lymphoblast proliferation. Proc. Natl. Acad. Sci. U.S.A. 113, 14121–14126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calo E., Wysocka J., Modification of enhancer chromatin: What, how, and why? Mol. Cell 49, 825–837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura H. et al., Global changes in Kaposi’s sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77, 4205–4220 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Core L. J., Waterfall J. J., Lis J. T., Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hutin S., Lee Y., Glaunsinger B. A., An RNA element in human interleukin 6 confers escape from degradation by the gammaherpesvirus SOX protein. J. Virol. 87, 4672–4682 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu J. et al., LANA binds to multiple active viral and cellular promoters and associates with the H3K4methyltransferase hSET1 complex. PLoS Pathog. 10, e1004240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H. S., Wikramasinghe P., Showe L., Lieberman P. M., Cohesins repress Kaposi’s sarcoma-associated herpesvirus immediate early gene transcription during latency. J. Virol. 86, 9454–9464 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou F. et al., Oncolytic reactivation of KSHV as a therapeutic approach for primary effusion lymphoma. Mol. Cancer Ther. 16, 2627–2638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hilton I. B. et al., The open chromatin landscape of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 87, 11831–11842 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thakore P. I. et al., Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delmore J. E. et al., BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tolani B., Gopalakrishnan R., Punj V., Matta H., Chaudhary P. M., Targeting Myc in KSHV-associated primary effusion lymphoma with BET bromodomain inhibitors. Oncogene 33, 2928–2937 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filippakopoulos P. et al., Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manzano M. et al., Gene essentiality landscape and druggable oncogenic dependencies in herpesviral primary effusion lymphoma. Nat. Commun. 9, 3263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forero A., McCormick K. D., Jenkins F. J., Sarkar S. N., Downregulation of IRF4 induces lytic reactivation of KSHV in primary effusion lymphoma cells. Virology 458-459, 4–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwata A. et al., Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF-IRF4 transcription factor complex. Nat. Immunol. 18, 563–572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee H. R. et al., Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor 4 (vIRF4) targets expression of cellular IRF4 and the Myc gene to facilitate lytic replication. J. Virol. 88, 2183–2194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin Q. et al., Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li M. et al., Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol. Cell. Biol. 20, 8254–8263 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacobs S. R., Damania B., The viral interferon regulatory factors of KSHV: Immunosuppressors or oncogenes? Front. Immunol. 2, 19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toth Z. et al., Negative elongation factor-mediated suppression of RNA polymerase II elongation of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. J. Virol. 86, 9696–9707 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clyde K., Glaunsinger B. A., Deep sequencing reveals direct targets of gammaherpesvirus-induced mRNA decay and suggests that multiple mechanisms govern cellular transcript escape. PLoS One 6, e19655 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsieh C. L. et al., Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. U.S.A. 111, 7319–7324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alvarez-Dominguez J. R., Knoll M., Gromatzky A. A., Lodish H. F., The super-enhancer-derived alncRNA-EC7/bloodlinc potentiates red blood cell development in trans. Cell Rep. 19, 2503–2514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heward J. A., Roux B. T., Lindsay M. A., Divergent signalling pathways regulate lipopolysaccharide-induced eRNA expression in human monocytic THP1 cells. FEBS Lett. 589, 396–406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li W. et al., Condensin I and II complexes license full estrogen receptor α-dependent enhancer activation. Mol. Cell 59, 188–202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S., Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stadhouders R. et al., Multiplexed chromosome conformation capture sequencing for rapid genome-scale high-resolution detection of long-range chromatin interactions. Nat. Protoc. 8, 509–524 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Li W. et al., Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park A., et al. , Global epigenomic analysis of primary effusion lymphoma identifies MYC super-enhancers and enhancer RNAs associated with KSHV infection. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147063. Deposited 16 March 2020.
- 62.Hu J., Yang Y., McIntyre L., Renne R., Genome-wide ChIP sequence analysis of KSHV-infected primary effusion lymphoma (PEL) cell line BCBL-1 and human umbilical vein endothelial TIVE-LTC cells. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM1265857. Accessed 1 August 2018.
- 63.Izumiya Y., Zhou F., Tepper C. G., Oncolytic reactivation of KSHV as a therapeutic approach for primary effusion lymphoma: High-resolution mapping of BRD4 binding sites in the KSHV genome. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM2769881. Accessed 1 August 2018.
- 64.Hu J., Yang Y., McIntyre L., Renne R., Genome-wide ChIP sequence analysis of KSHV-infected primary effusion lymphoma (PEL) cell line BCBL-1 and human umbilical vein endothelial TIVE-LTC cells. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM1265864. Accessed 1 August 2018.
- 65.Chen H. S., Wikramasinghe P., Showe L., Lieberman P. M., Cohesins repress KSHV immediate early gene transcription during latency. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM941710. Accessed 1 August 2018.
- 66.Chen H. S., Wikramasinghe P., Showe L., Lieberman P. M., Cohesins repress KSHV immediate early gene transcription during latency. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM941711. Accessed 1 August 2018.
- 67.Hilton I. B., Simon J. M., Davis I. J., Dittmer D. P., The open chromatin landscape of Kaposi's sarcoma-associated herpesvirus. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM1223899. Accessed 1 August 2018. [DOI] [PMC free article] [PubMed]
- 68.Papp B., Toth Z., Global host gene expression changes in KSHV+ PEL cells upon KSHV reactivation. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE123898. Accessed 1 June 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GRO-seq, 4C-seq, and H3K27ac ChIP-seq data were deposited in NCBI Gene Expression Omnibus (GEO) database under accession number GSE147063 (61). Previously reported ChIP-seq data (H3K4me3, BRD4, RNAPII, CTCF, SMC1), FAIRE-seq data, and RNA-seq data were used to support this study and are available in NCBI GEO database under accession numbers GSM1265857, GSM2769881, GSM1265864, GSM941710, GSM941711, and GSE123898, respectively (62–68). These datasets are cited at relevant places within the text.