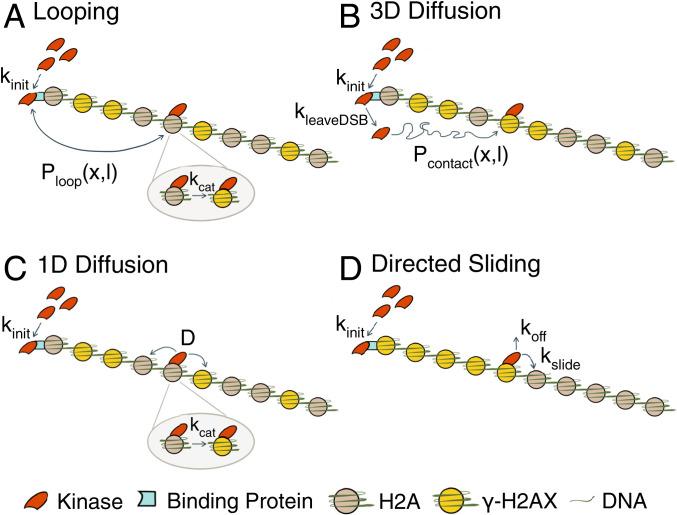
Fig. 3.
Schematic of phosphorylation spreading mechanisms. Schematics of (A) looping, (B) 3D diffusion, (C) 1D diffusion, and (D) directed sliding mechanisms. The kinases Mec1 and Tel1 (red) are recruited to the DSB at a rate kinit to RPA and MRX respectively (teal). For 1D diffusion and directed sliding, kinit comprises both the recruitment to and detachment from the DSB of the kinase to begin translocating along the chromatin. The kinase proceeds to phosphorylate H2A (gray) to form γ-H2AX (yellow) using one of the four mechanisms. (A) In looping, the kinase remains tethered at the DSB and forms a looped conformation with probability Ploop(x,l), where x = distance in kilobases of the target H2A from the break and l is the chromatin Kuhn length. Ploop(x,l) is formulated using a worm-like chain polymer model. The kinase phosphorylates H2A at a rate kcat after contact. (B) In 3D diffusion, the kinase transiently binds to the DSB where it is activated and released at a rate kleaveDSB. The activated kinase diffuses away from the DSB until it encounters and phosphorylates H2A. Pcontact(x,l) is the probability that the kinase comes into contact with an H2A located a distance x away from the break. (C) In 1D diffusion, the kinase is recruited to the DSB and proceeds to move along the chromatin with a 1D diffusion coefficient D. The kinase phosphorylates H2A that it comes into contact with at a rate kcat. (D) In directed sliding, the kinase moves unidirectionally away from the DSB along the chromatin. The kinase slides onto adjacent histones at a rate kslide and phosphorylates all H2As encountered until the kinase detaches from the chromatin at a rate koff.