Significance
Vertebrate retinal ganglion cells (RGCs) transmit all visual signals from the eye to the brain, are the pathogenic target in glaucoma, and require the Atoh7 competence factor to develop from multipotent progenitors. Atoh7 transcription is controlled by dual cis regulatory elements, including a remote shadow enhancer (SE). In humans, loss of the SE causes NCRNA disease, with congenital blindness due to optic nerve aplasia. We generated a mouse Atoh7 SE deletion model and analyzed its effects on transcription, retinal histology, and chromatin architecture. The mutant mice express 80% less Atoh7 mRNA but retain optic nerves, unlike NCRNA patients. By systematically varying dosage in a genotypic series, we show how the dual enhancers maintain robust plasticity during RGC genesis.
Keywords: glaucoma, shadow enhancer, optic disc area, optic nerve, human genetic disorders
Abstract
The retinal ganglion cell (RGC) competence factor ATOH7 is dynamically expressed during retinal histogenesis. ATOH7 transcription is controlled by a promoter-adjacent primary enhancer and a remote shadow enhancer (SE). Deletion of the ATOH7 human SE causes nonsyndromic congenital retinal nonattachment (NCRNA) disease, characterized by optic nerve aplasia and total blindness. We used genome editing to model NCRNA in mice. Deletion of the murine SE reduces Atoh7 messenger RNA (mRNA) fivefold but does not recapitulate optic nerve loss; however, SEdel/knockout (KO) trans heterozygotes have thin optic nerves. By analyzing Atoh7 mRNA and protein levels, RGC development and survival, and chromatin landscape effects, we show that the SE ensures robust Atoh7 transcriptional output. Combining SE deletion and KO and wild-type alleles in a genotypic series, we determined the amount of Atoh7 needed to produce a normal complement of adult RGCs, and the secondary consequences of graded reductions in Atoh7 dosage. Together, these data reveal the workings of an evolutionary fail-safe, a duplicate enhancer mechanism that is hard-wired in the machinery of vertebrate retinal ganglion cell genesis.
The vertebrate retina contains six major neuronal cell types—retinal ganglion cells (RGCs); horizontal, amacrine, and bipolar interneurons; and rod and cone photoreceptors—and Müller glia, which differentiate from a multipotent retinal progenitor cell (RPC) population in a stereotyped but overlapping temporal birth order, in response to intrinsic and extrinsic cues (1, 2). RGCs are the first-born retinal cell type in all species examined and actively regulate downstream histogenetic events. For example, RGCs secrete sonic hedgehog (SHH), which drives RPC proliferation, affecting the size of later born retinal cell populations, signals astrocytes, and controls morphogenesis of the optic stalk (3, 4). Later in development, RGC axons, which form the optic nerve, provide an anatomical path for astrocytes to enter the optic cup and promote development of definitive retinal vasculature (5–7). RGCs ultimately transmit all visual information from the eye to the brain, and their axons determine the bandwidth for this transfer. The birth and maintenance of RGCs is thus an important facet of retinal development and disease.
The ATOH7 (atonal homolog) transcription factor critically regulates RGC genesis by establishing a ganglion cell competence state in retinal progenitors during their terminal mitotic division (8, 9). This conserved proneural basic helix–loop–helix (bHLH) protein is transiently and dynamically expressed during vertebrate retinal histogenesis, in a spatiotemporal pattern that directly precedes the wave of RGC differentiation, from embryonic day 11 (E11) to postnatal day 0 (P0) in mice (10, 11). At the cellular level, mouse Atoh7 expression begins during the terminal S/G2 phase and ends as postmitotic daughter cells reach their final laminar position in the retina; however, the onset of Atoh7 expression within the last cell cycle is progressively delayed from E11 to P0 (8, 12–14). Analyses of mutant mice and zebrafish show ATOH7 is an essential, rate-limiting factor for ganglion cell genesis (15–19). Atoh7 mutants lack RGCs and optic nerves but retain all other cell types. Despite this selectivity, all seven major retinal cell types are represented in the Atoh7 lineage, such that only 11% of Atoh7+ cells adopt an RGC fate (8, 20). Moreover, Cre-lox lineage data show that only 55% of mature RGCs derive from Atoh7+ cells, suggesting that ATOH7 has additional nonautonomous effects (8).
Atoh7 transcription is regulated in part by conserved DNA elements (proximal and distal) located within 2.6 kilobases (kb) of the transcription start site (TSS). They harbor confirmed binding sites for PAX6, NEUROG2, and RBP-Jκ (CSL) transcription factors, but these features do not fully explain the dynamic pattern of Atoh7 expression (21–25).
Human nonsyndromic congenital retinal nonattachment (NCRNA) disease is characterized by optic nerve aplasia, with profound secondary retinovascular and other ocular defects (26, 27). The clinical phenotype overlaps autosomal recessive persistent hyperplastic primary vitreous (arPHPV) and vitreoretinal dystrophy, which are caused by point mutations in the ATOH7 coding sequence (18, 26, 28, 29), and findings in Atoh7 mutant mice (16, 17, 30). NCRNA is caused by a 6.5-kb deletion, extending from 19.2 to 25.7 kb upstream of the TSS. The deletion spans three conserved noncoding elements (CNEs), which together recapitulate the expression pattern of endogenous Atoh7 in transgenic mice and zebrafish (26). These remote CNEs share no obvious DNA sequence homology with proximal and distal CNEs located close to the TSS, which constitute the “primary” enhancer (PE), but their expression patterns appear identical. The NCRNA deletion thus removes a remote “shadow” enhancer (SE) that is vital for full ATOH7 expression and RGC genesis but also, paradoxically, redundant with the PE. Moreover, multiple genome-wide association studies (GWAS), in diverse human populations, suggest that ATOH7 is the major determinant of variation in optic disk size, a clinical indicator of RGC number—with peak logarithm-of-the-odds (LOD) scores directly overlying the SE (31–35). How do these dual enhancers coordinately regulate Atoh7 transcription? How do Atoh7 coding and SE regulatory mutations differ in phenotypic severity and developmental progression?
Duplicate regulatory elements have been described for several genes (36–39), including vertebrate retinal genes (40). Broadly speaking, PEs and SEs have distinct evolutionary origins but highly overlapping spatiotemporal domains, and they control transcription of the same gene. Their names do not indicate hierarchy or dominance; PEs are simply located closer to the TSS (41, 42). In some circumstances, one enhancer (PE or SE) can fully initiate and maintain gene expression by itself (43, 44)—yet both have been positively selected during evolution. In the case of ATOH7, the PEs and SEs may have complementary or synergistic roles that improve fitness. Because ATOH7 specifies the first retinal cell type, these enhancers control the onset of neurogenesis in the vertebrate eye.
To understand dual ATOH7 enhancer function and NCRNA pathogenesis, we generated SE deletion (Atoh7SE) mice via CRISPR/Cas9 editing. By combining SE deletion and knockout (KO) alleles, we created a genotypic series with six graded levels of Atoh7 expression and quantitatively characterized effects on RGC genesis and retinal development. The SE deletion profoundly reduced Atoh7 transcription but did not alter its spatial or temporal pattern, or the architecture of surrounding chromatin. The number of adult RGCs was correlated with Atoh7 mRNA abundance following a saturation curve, and there were secondary defects in optic nerve head (ONH) morphology, axon fasciculation, and retinal vasculature. However, unlike humans with NCRNA disease, the optic nerves of homozygous Atoh7SE mice were grossly normal. These dual modular enhancers can act additively to ensure robustness of expression, allowing developmental programs and regulatory networks to resist environmental or genetic perturbations.
Results
The SE Regulates Atoh7 Transcription.
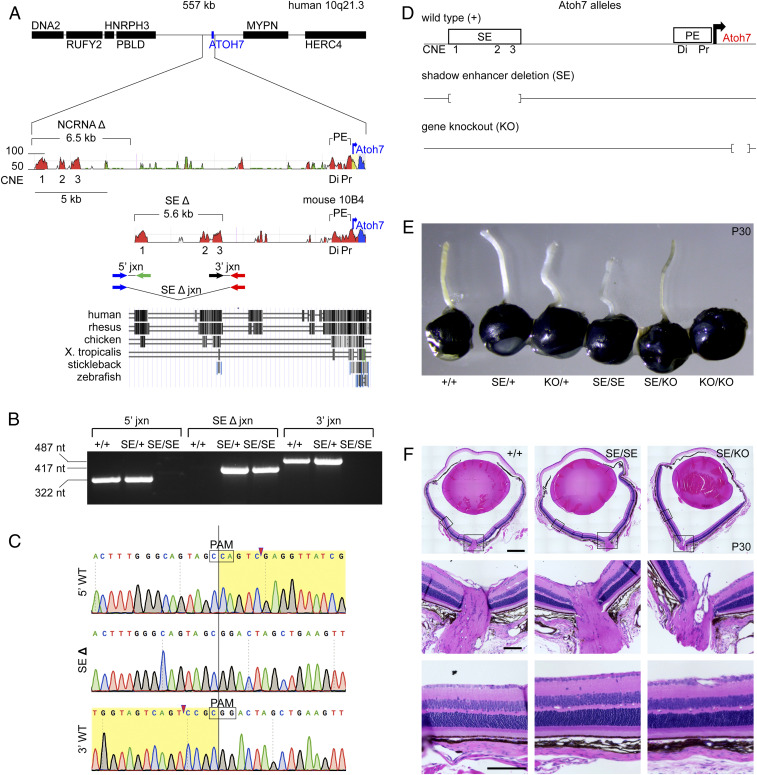
To model NCRNA, we deleted 5.6 kb of mouse chromosome 10 spanning the Atoh7 SE (Fig. 1A). We generated three Atoh7 SE deletion alleles, with similar homozygous eye phenotypes, and chose one for further investigation (Fig. 1 B and C and SI Appendix, Fig. S1). Unlike NCRNA patients and Atoh7 KO animal models, the SE/SE mice had intact optic nerves (Fig. 1E). However, the optic nerves of trans heterozygous (SE/KO) eyes were noticeably thinner than wild-type (WT), with no change in retinal morphology (Fig. 1 E and F), suggesting that loss of the SE does impair Atoh7 expression and RGC genesis.
Fig. 1.
Targeted deletion of the murine Atoh7 SE. (A) Human and mouse Atoh7 locus maps. (Top) VISTA plots (114) show conserved noncoding elements within SEs (CNE1-3) and PEs (Di, Pr). Human NCRNA and mouse SE deletions (brackets) and transcription start sites (blue arrows) are indicated. Sequence identity (50 to 100%) is plotted for a 50-bp moving window. DNA segments are identified as intergenic (red), repetitive (green), coding (blue), or UTR (yellow). Primers used to amplify SE deletion (Δ) junction and WT endpoints are indicated below the mouse VISTA plot. (Bottom) Multiz species alignment, adapted from the University of California Santa Cruz (UCSC) browser (build mm10). (B) Agarose gel showing diagnostic PCRs from +/+, SE/+, and SE/SE genomic DNA. (C) Sanger chromatograms of PCR products spanning the SEΔ junction and WT endpoints. Deleted nucleotides (yellow highlight), Cas9 cut sites (arrowheads), and protospacer adjacent motifs (PAMs) are indicated. (D) Schema of Atoh7 alleles. (E) Adult eyes from Atoh7 genotypic series. The optic nerves are thin in SE/KO eyes (with 56.6 ± 10.6% of +/+ cross-sectional area, SI Appendix, Table S4) and absent in KO/KO eyes. (F) Histology of adult eyes (hematoxylin/eosin) with similar retinal morphology. X. tropicalis, Xenopus tropicalis; nt, nucleotides. (Scale bars: Top, 500 µm; and Middle and Bottom, 100 µm.)
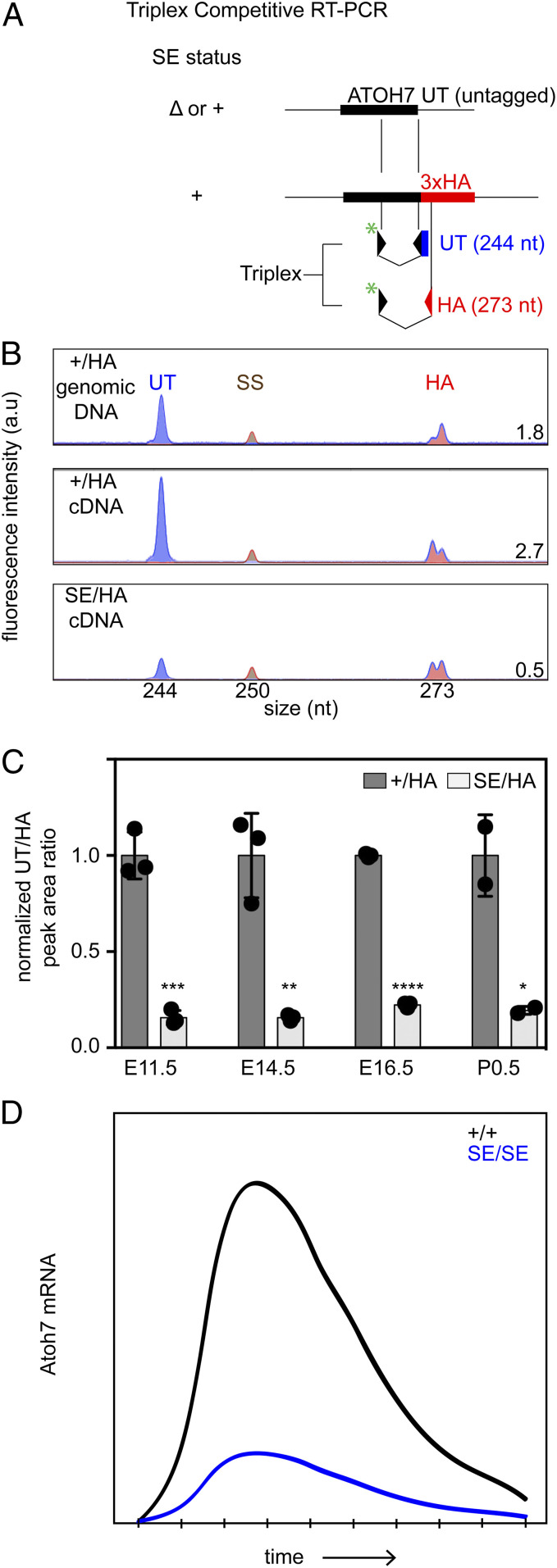
In principle, enhancers can regulate the level, timing, and/or spatial pattern of gene expression. To determine how SE deletion affects the level and timing of transcription in cis, we measured the relative abundance of Atoh7 messenger RNA (mRNA) transcripts in a triplex competitive reverse transcriptase (RT)-PCR experiment (Fig. 2). We compared the molar ratio of PCR products from littermate +/HA (hemagglutinin) and SE/HA retinas at the onset (E11.5), peak (E14.5), and offset (E16.5, P0.5) of Atoh7 expression (8) using the trans Atoh7HA (HA epitope tag) knock-in allele (45) as an internal reference to calculate the ratio of mRNAs from untagged (UT) WT and SE alleles (Fig. 2A). The HA epitope is coextensive with the endogenous Atoh7 protein in immunostained HA/+ retinas (ruling out allelic exclusion) (14) and has no detectable effect on RGC genesis (13, 45). If the SE deletion caused the temporal pattern to shift—delaying the onset of Atoh7 expression during development, for example—then the ratio of mRNAs originating from SE and HA alleles would be significantly reduced at early time points (E11.5) only. However, in these assays, the SE deletion caused a decrease in Atoh7 expression at every time point, by a similar magnitude (6.4-, 6.4-, 4.5-, and 5.2-fold at E11.5, E14.5, E16.5, and P0.5, respectively). The SE deletion thus uniformly reduces the level of Atoh7 expression four- to sixfold in cis, but does not alter the timing of transcription.
Fig. 2.
The SE regulates Atoh7 mRNA abundance. (A) Triplex RT-PCR strategy to quantify Atoh7 mRNAs in +/HA and SE/HA retinas. The FAM (*)-labeled 5′ primer (black arrowhead) recognizes both alleles; unlabeled 3′ primers are specific for UT (black/blue) or 3xHA knock-in (red) alleles. (B) Capillary electrophoresis profiles of PCR products from +/HA and SE/HA cDNA, and HA/+ genomic DNA. The average ratio of UT (244 nt) and HA (273 nt) cDNA peak areas, at the right-hand corner of each trace, reflects the relative abundance of allelic transcripts. The gDNA trace (UT/HA product ratio = 1.8 ± 0.11, n = 3) was included as a PCR efficiency control, with a 1.0 fixed molar template ratio. Atoh7 gene and cDNA are colinear with no introns. SS, 250-nt ROX size standard. (C) UT/HA cDNA peak area ratios are plotted at four ages, normalized to +/HA cDNA controls. The ratio of Atoh7 mRNAs transcribed from SEΔ versus WT alleles is 0.16 ± 0.02 at E11.5, 0.16 ± 0.009 at E14.5, 0.22 ± 0.007 at E16.5, and 0.20 ± 0.015 at P0.5. (D) Temporal profile of Atoh7 mRNA extrapolated from qPCR (8) and triplex RT-PCR data. The SE deletion uniformly reduces Atoh7 expression at all time points. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
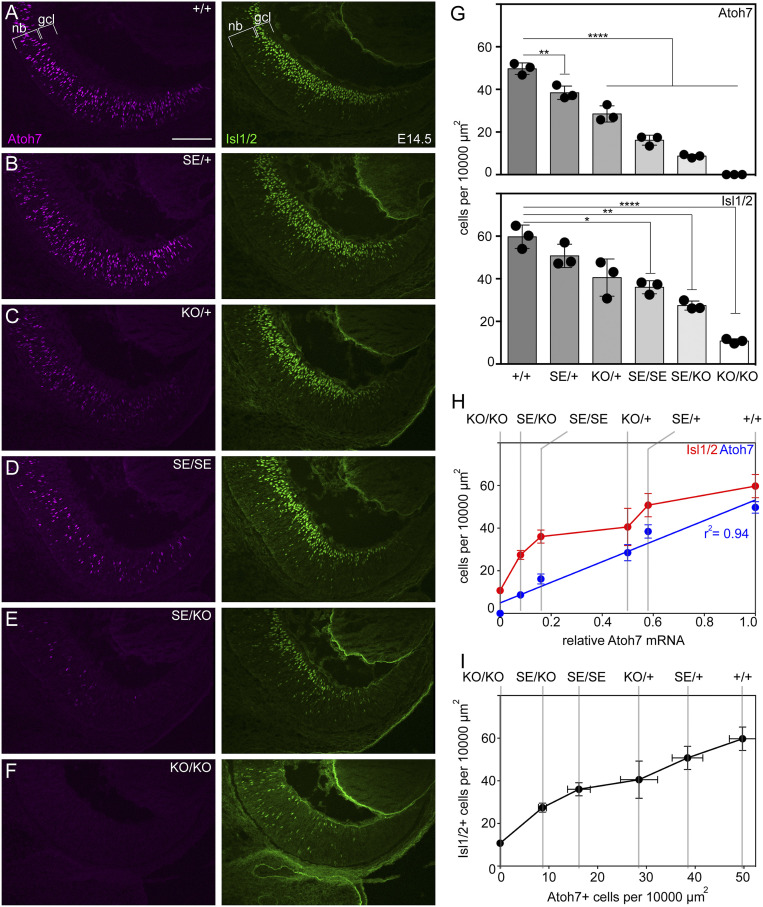
SE Mutants Have Fewer Atoh7+ and Isl1/2+ Cells.
To evaluate how the SE deletion affects the level of Atoh7 protein and distribution of Atoh7+ cells, we immunostained midaxial retina sections at E14.5 (peak expression) (Fig. 3) with a validated antibody (14) and determined planimetric cell densities. This value broadly reflects the size of the RGC-competent progenitor pool. We observed significantly fewer Atoh7+ cells per 10,000-µm2 retinal area for all five mutant genotypes in our series, compared to WT, but there were no differences in the overall retinal staining pattern (Fig. 3) or midaxial area (SI Appendix, Table S1). Across the genotypic series, the number of Atoh7+ cells was well correlated with the abundance of Atoh7 mRNA (Fig. 3H) (r2 = 0.94, P < 0.01, df = 4), using 1.00, 0.16, and 0.00 as normalized expression values for WT, SE, and KO alleles, respectively, based on triplex RT-PCR data.
Fig. 3.
Abundance of Atoh7+ and Isl1/2+ cells decreases across the Atoh7 genotypic series. (A–F) At E14.5, Atoh7+ cells (magenta) reside within the neuroblast layer (nb) layer while Isl1/2+ cells (green) are located in the nb and ganglion cell layer (gcl). KO/KO retinas lack Atoh7 immunoreactivity. (G) Atoh7+ and Isl1/2+ cell counts per 10,000 µm2. Each mutant genotype has fewer Atoh7+ cells than WT (+/+) and most have fewer Isl1/2+ cells. (H) In bivariate plots, the number of Atoh7+ and Isl1/2+ cells is directly related to Atoh7 mRNA abundance (for Atoh7+ cells, linear regression P < 0.01, r2 = 0.94, df = 4). Atoh7 mRNA values were calculated from triplex RT-PCR data as 1.00 (+/+), 0.58 (SE/+), 0.50 (KO/+), 0.16 (SE/SE), 0.08 (SE/KO), and 0.00 (KO/KO). (I) The numbers of Atoh7+ and Isl1/2+ cells are strongly correlated across the genotypic series. *P < 0.05, **P < 0.01 and ****P < 0.0001. (Scale bar: 100 µm.)
We then performed a comparable analysis for LIM homeobox factors Islet-1 and 2 using the same E14.5 retinas, which were costained with anti-Isl1/2 (Fig. 3). During normal development, retinal progenitors oscillate radially within the neuroepithelium via interkinetic nuclear migration (IKNM), coupled to the cell cycle (46). Following terminal mitosis at the retinal apex, nascent RGCs migrate basally. At day E14.5, Isl1 and Isl2 are expressed in differentiating RGCs and starburst amacrine cells (47) in the inner neuroblast layer and overlap with Atoh7 in a narrow postmitotic transition zone. As nascent RGCs traverse this zone, they down-regulate Atoh7 and up-regulate Isl1/2 (SI Appendix, Fig. S2). Isl1 expression is downstream of Atoh7 whereas Isl2 is immediately downstream of Isl1 (48–50); both are continuously expressed in mature RGCs. We observed a similar decrease in the planimetric density of Isl1/2+ cells across the Atoh7 genotypic series. The presence of Isl1/2+ cells in Atoh7 KO/KO retinas is consistent with previous results showing the existence of an alternate, independent mode of Isl1 regulation during RGC genesis (49). Nonetheless, the overall number of Isl1/2+ cells was correlated with Atoh7 mRNA levels (Fig. 3H) and the number of Atoh7+ cells (Fig. 3I), as expected. Similar decreases in Brn3+ (Pou4f) and Rbpms+ (multiply spliced RNA binding protein) cells were observed in Atoh7 SE/SE and SE/KO E14.5 retinas (SI Appendix, Fig. S3). The concordance between Atoh7 and Isl1/2 (ratio of double-positive to all positive cells) also decreased across genotypes but declined most abruptly between KO/+ (8.0 ± 1.7%) and SE/SE (3.0 ± 0.6%) retinas (SI Appendix, Fig. S2), reflecting a shorter temporal overlap between Atoh7 and its target, and potentially weaker transcriptional activation as Atoh7 protein levels are reduced. Loss of the SE thus causes a decrease in Atoh7 mRNA and Atoh7+ cells, and a corresponding decrease in Isl1/2+, Brn3+, and Rbpms+ cells, as indicators of RGC genesis.
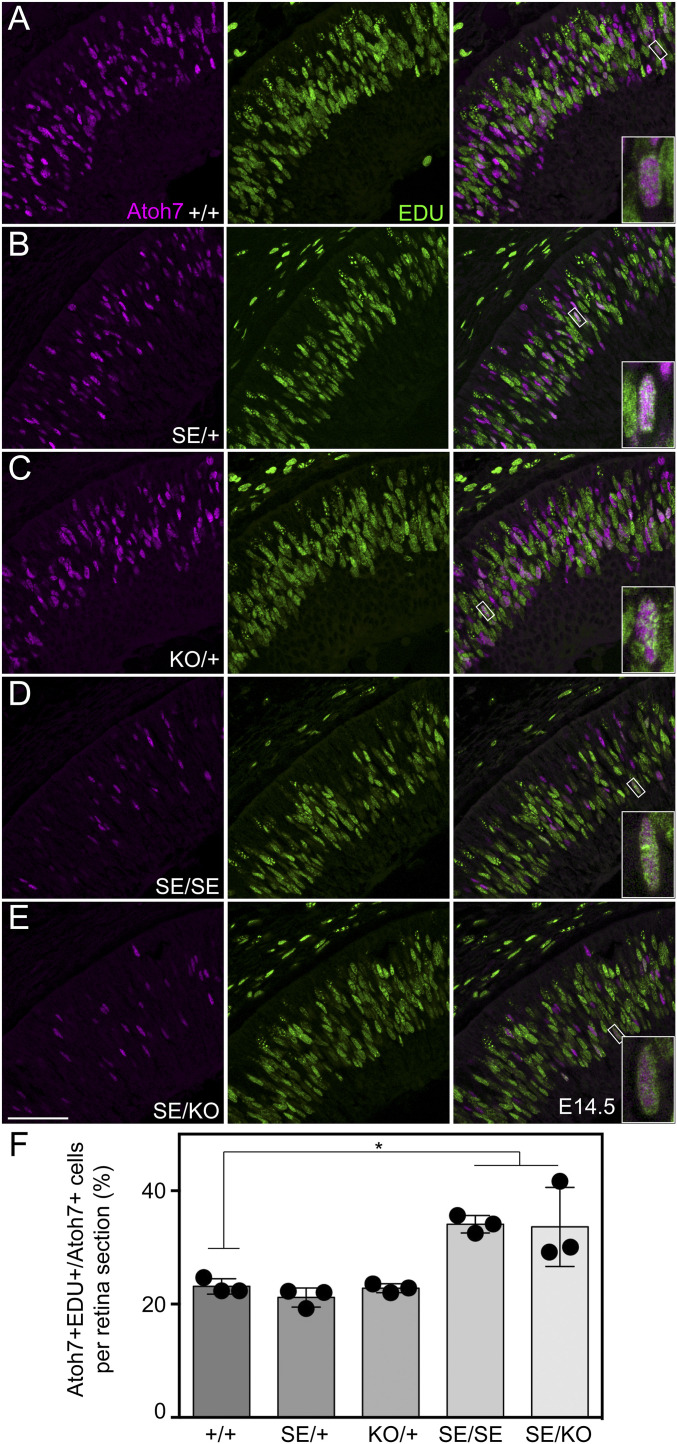
Altered Timing of Atoh7 Expression within the Terminal Cell Cycle.
At the onset of retinal neurogenesis (E11.5), a subset of progenitor cells experience a burst of Atoh7 transcription during terminal S-phase (13, 50, 51). As development proceeds (>E13.5) and RPC cycles lengthen (52–54), this transient Atoh7 burst is progressively delayed, into terminal G2/M and G0 phases (8, 51). While perdurant SE and PE reporter transgenes have coextensive spatiotemporal and cellular patterns (26) and the overall timing of Atoh7 expression is broadly preserved at the tissue level in SE mutants, in stochastically phased RPCs (Fig. 2), these dual enhancers may act differently on a cellular scale, controlling distinct temporal features of Atoh7 transcription within the last cycle. To test this possibility, we determined the S-phase fraction of Atoh7+ cells by EdU (5-ethynyl-2′-deoxyuridine) pulse labeling (Fig. 4). If Atoh7 activation were delayed in SE mutants, this value is expected to decrease. Instead, the S-phase fraction was significantly elevated in E14.5 SE/SE (34.1 ± 1.6%) and SE/KO (33.6 ± 7.0%) retinas compared to WT (23.1 ± 1.3%), suggesting that both enhancers (or the SE alone) may be needed to fully sustain the burst of Atoh7 transcription through the terminal cell cycle. At the same time, there was no change in the M-phase fraction, determined by PH3 (phosphohistone) staining (SI Appendix, Table S2). The accelerated decline in postmitotic Atoh7 expression in SE/SE and SE/KO retinas is supported by reduced Isl1/2 concordance (SI Appendix, Fig. S2).
Fig. 4.
Atoh7 declines faster in terminal S-phase in SE deletion mutants. (A–E) Distribution of Atoh7+ and EdU+ cells in E14.5 retinas. Double-positive cells, which have initiated Atoh7 expression during terminal S-phase (90-min EdU pulse), are present in all genotypes (Inset). (F) A greater fraction of Atoh7+ cells are EdU+ in SE/SE and SE/KO retinas compared to +/+. *P < 0.05. (Scale bar: 50 µm.)
Cone Genesis and Apoptosis in SE Mutant Retinas.
Beyond the absence of RGCs and optic nerves, adult Atoh7 knockout mice have thinner retinas, with a deficiency of later-born cell types that arises from decreased RPC proliferation (30). The cone-to-rod ratio is increased accordingly. In mutant embryos, retinal cell death is also broadly elevated (12, 16, 20). To evaluate cone genesis and apoptosis in SE mutants, we stained E14.5 retinas with thyroid hormone receptor beta (Thrβ2) and cleaved polyADP ribose polymerase (c-PARP) antibodies, respectively (SI Appendix, Fig. S4). Apoptotic cells were more abundant in KO/KO retinas, but no other genotypes, and there was no major change in cones.
Postnatal Atoh7 SE Mutants Have Fewer RGCs.
In a previous study, we showed that a 50% reduction in Atoh7 dosage impairs RGC genesis but does not alter the final number of RGCs, by comparing optic nerve axons in neonatal and adult Atoh7 +/+ and KO/+ mice (19). The developing nervous system thus compensates for defective RGC genesis by decreasing the extent of neonatal RGC culling (55, 56), a normal process that matches the number of RGCs to postsynaptic targets in the brain. To test whether similar mechanisms operate in Atoh7 SE mutants, we first compared cross-sectional areas of P1 optic nerves in SE/SE and SE/KO animals, which were significantly smaller than WT (SI Appendix, Fig. S5 and Table S3). At P1, darkly stained nuclei, presumed to be glial, were also prominent in thin sections. We observed fewer nuclei in mutant optic nerves, but no change in density (SI Appendix, Table S3).
At P30, after RGC culling, the cross-sectional area of SE/SE optic nerves was reduced but not significantly different from WT. In contrast, SE/KO nerves were significantly thinner at both ages, with a greater difference at P1 than at P30 (SI Appendix, Fig. S5 and Table S4). We further analyzed P30 optic nerves by electron microscopy (EM) to determine the number of myelinated axons, which corresponds directly to the number of RGCs. The axon density was similar across genotypes, but, because SE/KO optic nerves were smaller, they had significantly fewer axons (SI Appendix, Table S4). Reduced Atoh7 dosage thus restricts RGC genesis in the NCRNA animal model. In SE/SE mice, this defect is largely compensated by a reduction in postnatal RGC culling, since the range of these effects is similar, approximately twofold (Fig. 3) (56). However, in SE/KO mice, the deficiency in RGC genesis (fivefold) exceeds the buffering capacity of the culling mechanism so the number of surviving RGCs in adults is decreased (two- to threefold).
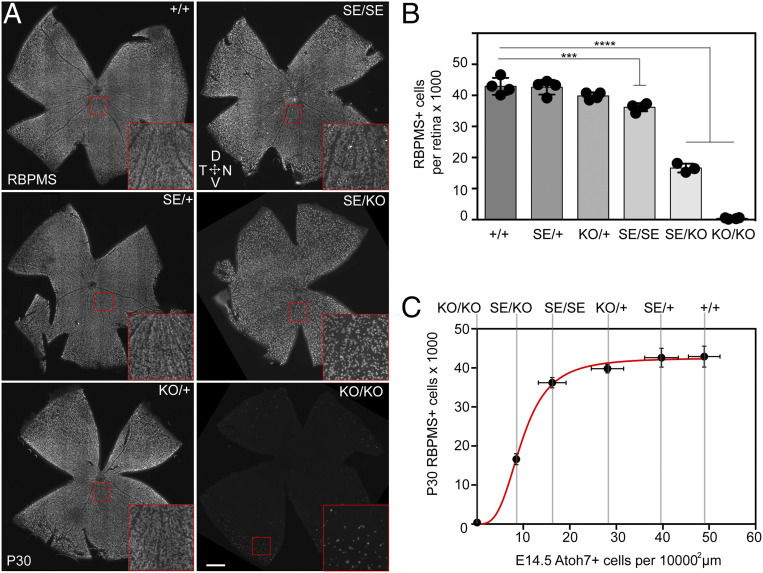
Since RGC myelination continues until postnatal week 16 in mice (57), axon counts may, in principle, underestimate the true number ganglion cells in Atoh7 SE mutants. We therefore determined the total number of Rbpms-immunoreactive RGCs in P30 retinal flatmounts (58) using an automated cell counting algorithm (SI Appendix, Fig. S6). This allowed us to quantify RGCs with greater precision across the full Atoh7 genotypic series (Fig. 5 and SI Appendix, Table S5). Our WT C57BL/6 data match those obtained via other automated counting approaches (59, 60). Consistent with previous analyses (19), there was no significant difference in the number or density of RGCs in adult Atoh7 KO/+ and SE/+ retinas compared to WT, and KO/KO retinas had scant RGCs. However, both SE/KO and SE/SE retinas had significantly fewer RGCs. The distribution of RGCs was similar in all genotypes, with lowest density in the dorsal periphery and highest density in a central peripapillary stripe (SI Appendix, Fig. S7), as previously noted in mice (61) and rats (62). The number of adult RGCs varied with the size of the Atoh7+ progenitor pool, following a saturation curve (Fig. 5C).
Fig. 5.
Fewer RGCs in adult SE mutants. (A) P30 retinal flatmounts immunostained with Rbpms antisera. SE/KO retinas have noticeably fewer Rbpms+ RGCs while KO/KO retinas have essentially none. Insets show 4×-magnified views. (B) Automated cell count data showing fewer total Rbpms+ RGCs in SE/SE, SE/KO, and KO/KO retinas than in +/+ retinas. (C) Plot showing how adult RGC counts vary with size of the RGC-competent progenitor pool, indicated by the planimetric density of E14.5 Atoh7+ cells. The curve rises steeply but flattens sharply for Atoh7+ cell densities >30% of WT. ***P < 0.001 and ****P < 0.0001. (Scale bar: 500 µm.)
Secondary Effects in SE Mutant Retinas.
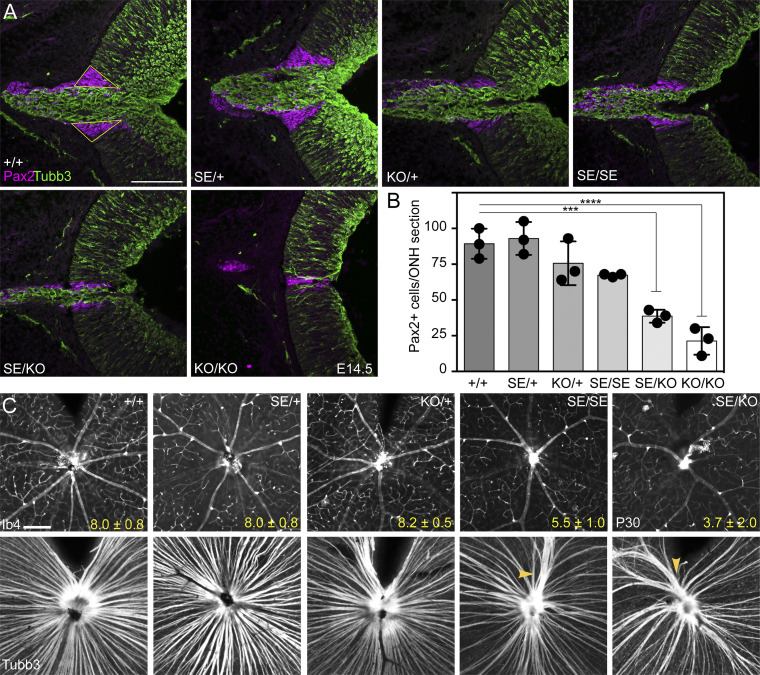
As RGC axons exit the eye and extend toward the diencephalon, they pass through the ONH, a discrete structure in the ventral posterior retina where the choroid fissure closes and hyaloid blood vessels enter (63). To investigate this structure and the glial deficiency in P1 mutant optic nerves (SI Appendix, Fig. S5), we counted astrocyte precursor cells (APCs) in the E14.5 ONH labeled with Pax2 antisera. These cells are packed in a wedge-shaped torus at the posterior pole of the optic cup, where it joins the optic stalk (Fig. 6 A and B), and differ in their properties from Pax2+ cells located inside the stalk (63–66). There were fewer ONH Pax2+ cells in SE/KO and KO/KO mice compared to WT, with smaller reductions in SE/SE and KO/+ mice. Thus, like other phenotypes that depend on RGCs, ONH development is impaired in Atoh7 mutants, with increasing severity across the genotypic series.
Fig. 6.
Secondary effects associated with graded loss of Atoh7 expression. (A) Pax2+ cells (magenta) delimit the ONH in an E14.5 mutant series. The intraretinal ONH domain (yellow outline in +/+ retina) does not include the stalk. (B) Comparison of Pax2+ ONH cells per meridional section across Atoh7 genotypes. The abundance of Pax2+ APCs is correlated with the number of Atoh7+ progenitors. SE/KO retinas have significantly fewer Pax2+ cells than +/+ retinas. (C) P30 retinal flatmounts showing surface vasculature (Ib4 lectin) and RGC axons (Tubb3). Fewer major blood vessels emanate from the ONH in SE/SE and SE/KO retinas compared to WT (number ± SD, n = 3). Axon fasciculation is abnormal in SE/SE and SE/KO retinas, with large aggregates in the central retina (yellow arrowheads) adjacent to the ONH. ***P < 0.001 and ****P < 0.0001. (Scale bar: 100 µm.)
Current models for retinal vasculogenesis highlight the importance of RGC axons, which haptotactically guide optic stalk astrocytes into the eye, where they direct endothelial cell migration and blood vessel development and SHH secreted by RGCs, which initiates this process (5, 6). These models are based on the major secondary vasculature defects associated with RGC agenesis in Atoh7 mutant animals and humans with PHPV or NCRNA disease (5, 16, 18, 26). In each case, intravitreal fetal (hyaloid) vessels persist, and the adult retinal vasculature does not develop. Vascular endothelial cells (VECs) fail to enter the eye from the ONH, due to astrocyte migration and polarization defects in the absence of ganglion cells (6). In WT mice, the influx of VECs occurs postnatally, after RGC genesis is complete. Neither astrocytes nor VECs express Atoh7.
To determine how graded reductions in Atoh7 dosage and RGC density alter retinal vasculature and the formation of axon bundles, we examined flatmounts costained for Rbpms (RGC soma), Ib4-lectin (vessels), and Tubb3 (neural tubulin β3, axons). There were two noteworthy changes (Fig. 6C). First, the mean number of major vessels emanating from the ONH was significantly reduced in SE/SE and SE/KO retinas compared to WT, SE/+, and KO/+. However, despite having fewer primary vessels and 61% fewer RGCs (SE/KO), these mutant retinas were fully vascularized by smaller arterioles and veins. A potentially similar decrease in retinovascular branching points has been noted in children with optic nerve hypoplasia (67). Second, axon fasciculation was abnormal in SE/SE and SE/KO retinas. In both genotypes, we observed large bundles of Tubb3+ fascicles and a unique pattern of axon tracts converging on the ONH, unlike WT retinas (Fig. 6C). These anatomical findings were supported by in vivo phase variance optical coherence tomography (pv-OCT) of the nerve fiber layer (NFL) and retinal vasculature in Atoh7 SE/SE and SE/KO mice (SI Appendix, Fig. S8). Our stepwise genotypic data thus suggest that a threshold number of embryonic RGCs—reflecting 20 to 30% of WT Atoh7 dosage in mice—are needed for normal vascular development and axon fasciculation.
Atoh7 Chromatin Accessibility in SE Mutant Retinas.
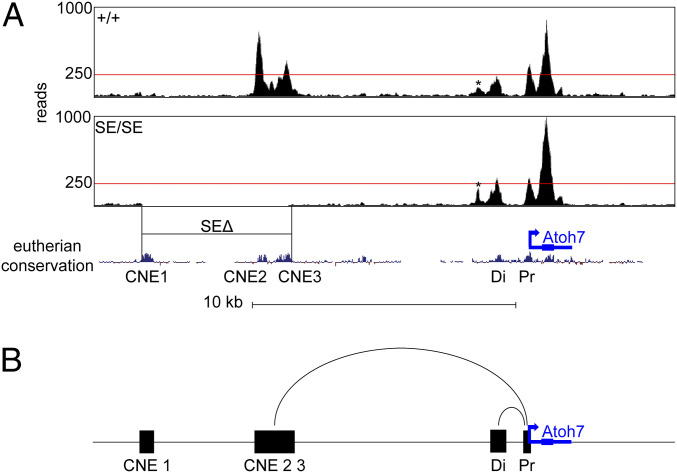
To evaluate Atoh7 chromatin architecture, we compared E14.5 retinas from WT and SE/SE animals using assay for transposase-accessible chromatin with ATAC-seq (assay for transposase-accessible chromatin using sequencing) methods (68). As expected, Atoh7 SE and PE had signatures of open chromatin (Fig. 7A). The discrete open segments in these profiles correspond to known CNEs with two exceptions—a small open region 0.7 kb upstream from the distal (Di) PE element, which is not conserved, and SE element CNE1, which was not accessible above background. Based on read density, CNE2 appears to be most accessible, with 854 peak reads per nucleotide, followed by CNE3, Pr (proximal), and Di. Interestingly, CNE2 contains the lead GWAS single-nucleotide polymorphism for optic disk size in humans, rs3858145 (26, 31, 33), and shows greater accessibility than the distal CNE, which contains binding sites for established regulators of Atoh7 transcription (21–23, 69, 70). In SE/SE mutants, open signatures for CNE2 and CNE3 were missing, as expected, but accessibility of the surrounding chromatin was unchanged. These findings are reminiscent of ATAC-seq data from mice with α-globin enhancer deletions (71). In that study, individual Hba enhancers were shown to influence transcription independently, in an additive manner, with single-element deletions causing only slight changes in the accessibility of surrounding chromatin.
Fig. 7.
Open chromatin signatures surrounding Atoh7. (A) (A) ATAC-seq profiles of +/+ and SE/SE mutant E14.5 retinas across the Atoh7 locus. The total number of aligned reads per nucleotide are shown as bigWig tracks in the UCSC mm10 genome browser, with 250 reads indicated (red line). Each plot reflects pooled data from three embryo replicates (six retinas, >69 million reads). Open chromatin peaks are apparent over CNE2, CNE3, Di, Pr, a small nonconserved element (*) and the Atoh7gene body. No reads were mapped to CNE2 or CNE3 in SE/SE chromatin; otherwise, the ATAC-seq profiles are similar. The open chromatin status and accessibility of PE does not depend on SE. (B) Proposed mechanism for Atoh7 regulation by SE and PE during retinal histogenesis. SE (CNE2/3 elements) and PE (distal element) loop independently with the Atoh7 promoter to activate transcription and are functionally redundant. Each modular enhancer is sufficient to control spatiotemporal expression, but they act additively to increase activity.
Likewise, our findings suggest that the Atoh7 SE does not broadly regulate chromatin access across the locus. Instead, the SE and distal PE appear to work as separate cis regulatory modules to independently drive Atoh7 transcription (Fig. 7B). By itself, the PE can activate low levels of expression, which are sufficient for RGC genesis in mice. Although Isl1 and Pou4f2 are downstream of Atoh7, neither gene appears to be a direct transcriptional target (72), and their ATAC-seq profiles were unchanged in SE/SE retinas (SI Appendix, Fig. S9).
Discussion
Atoh7 SE Deletion Mice Do Not Model NCRNA Disease.
Murine Atoh7 mutations differ from NCRNA disease in two important ways. First, as shown here, the SE deletion reduces but does not block RGC genesis, so mutant mice retain optic nerves, whereas, in NCRNA disease, there is no radiological evidence for optic nerve remnants, chiasm, or tracts (26). While NCRNA deletion effects on human ATOH7 transcription are unknown, the relative contributions of SE and PE may differ between species. Alternatively, the fraction of Atoh7+ progenitors that adopt ganglion cell fates may differ radically between primates and rodents. In mice, only 11% of Atoh7+ cells develop into RGCs, but each is competent to do so (8, 20), providing a buffer for the embryo from environmental or genetic perturbations. Indeed, as we reduced Atoh7 activity and the size of the RGC-competent progenitor pool (Fig. 3H), the number of adult RGCs did not decline dramatically until these values fell below 20% of WT (Fig. 5C). Presumably, the fraction of Atoh7+ RPCs acquiring RGC fates was increased. However, if most primate Atoh7+ cells were intrinsically biased toward RGC fates, a fivefold decrease in activity would severely limit ganglion cell development. The developmental plasticity of Atoh7+ cells may thus be greater in mice than humans. Likewise, nonautonomous Atoh7 effects on RGC genesis (8) may differ between species. Finally, RGC culling provides a basis to compensate for RGC deficiency and was diminished in SE homozygotes (SI Appendix, Fig. S5). The culled fraction, however, is similar in primates, rodents, and other vertebrates, roughly 50 to 70% (73–76), so cannot explain the huge difference in phenotypic severity between human and mouse SE deletions.
Second, in Atoh7-null mice, the hyaloid vasculature persists in the vitreous and invades the retina but does not proliferate excessively (5, 6, 77); however, in NCRNA disease, there is a dense profusion of retrolental fetal vessels, which fill the vitreous and retract, causing bilateral retinal detachments at birth. Indeed, while most anatomical features of the eye scale with size, cell dimensions and the physics of O2 diffusion do not. Since eye globe volumes are >400× larger in humans than mice, and RGCs are required for adult retinal blood vessels to develop, the magnitude of retinal hypoxia and vigor of the secondary neovascular response are expected to be much greater in NCRNA disease. Likewise, RGC axons provide anatomical support to stabilize the posterior globe. In the larger human eye, exposed to greater biomechanical force (78, 79), the retina may detach more readily than it does in mice when this structure is missing, severing any remaining RGC axons. While these secondary effects may contribute to NCRNA disease pathology, our data suggest impaired RGC fate specification is the major primary defect.
RGC Genesis Depends Strictly on Atoh7 Activity.
Using the hypomorphic SE allele, we extended previous analysis (19) to define Atoh7 dosage thresholds for RGC phenotypes. Two findings are striking. First, the abundance of Atoh7+ and Isl1/2+ cells depends directly on Atoh7 mRNA (Fig. 3H), which is uniformly decreased by the SE deletion at all ages (Fig. 2). The transient burst of Atoh7 appears necessary to kindle transcription of Isl1 and Pou4f2, which cooperatively instruct progenitors to differentiate as RGCs and positively regulate their own expression (48–50, 80, 81). Second, the number of mature RGCs (P30) is relatively stable until the Atoh7+ progenitor pool is diminished by >70% (Fig. 5C). Adult +/+, SE/+, and KO/+ mice have a similar number of RGCs, and SE/SE mice have only 16% fewer RGCs despite having 84% less Atoh7 mRNA. Indeed, widespread transgenic expression of Atoh7 in >80% of retinal progenitors during terminal mitosis, and continuously thereafter in Crx lineal descendants (photoreceptor and bipolar cells), does not increase the number of RGCs (82).
Timing of Atoh7 expression is also critical. Rescue of RGC genesis in Tg(Crx>Atoh7-ires-Cre); Atoh7 KO/KO mice is partial and restricted to progenitors that exit mitosis during the normal temporal envelope for RGC births (82). In this respect, the narrow time window for Atoh7 transcription, mediated by dual enhancers, is reminiscent of Drosophila atonal (83) and Sry, the HMG-box testis-determining factor. Sry is transiently expressed in bipotential gonads where it coactivates Sox9 transcription and initiates male sexual differentiation (84). When timing of the Sry pulse is shifted, even by 6 h, testis development is severely disrupted (85–87). Accordingly, dual Atoh7 enhancers may be needed as a fail-safe to ensure robust mRNA transcription during a short time interval. Despite these constraints, the period of RGC competence and Atoh7 expression can be prolonged under certain circumstances, leading to a marked increase in ganglion cells (88–90).
Decreased Atoh7 Activity Causes Secondary Malformations.
Loss of RGCs in Atoh7 mutants triggers a chain of downstream effects involving the ONH, retinal astrocytes, and vasculature. Fewer Pax2+ glia are present in ONH domains of Atoh7 SE/SE, SE/KO, and KO/KO mice (Fig. 6 A and B). These Pax2+ APCs enter the optic cup and give rise to A1 astrocytes, which are needed to establish the retinal vasculature (64, 65). Their migration depends on SHH signaling from pioneer RGC axons (3, 4, 66). Reduced ONH dimensions may also physically restrict migration of astrocytes and VECs into the retina.
One unique phenotype in the Atoh7 series is an alteration in axon fasciculation, with coarse bundling of axons converging on the ONH in SE/SE and SE/KO retinas (Fig. 6C). Hyperfasciculation may be mediated by reduced expression of downstream cell adhesion molecules, such as L1CAM (91), but has not been reported in Pou4f1-3, Isl1, Isl2, Sox4, or Sox11 mutants, which have comparable reductions in RGC abundance (48–50, 92). However, Pou4f2 and Isl1 mutants do exhibit axon pathfinding defects at the chiasm (49). Intraretinal axon misrouting has been noted in Bmp7 mutants, which lack an ONH (63), and in Tg(Crx>Atoh7-ires-Cre); Atoh7 KO/KO mice, with delayed RGC genesis (82).
The Atoh7 SE Does Not Control Access to Surrounding Chromatin.
CNEs often indicate the position of enhancers and, when active, exhibit specific histone acetylation (H3K27ac) and methylation (H3K4me1) marks, p300 enrichment, and features of open chromatin (42, 93, 94). The Atoh7 SE is structurally and functionally conserved; as such, the human SE drives reporter expression appropriately in transgenic mice and zebrafish (26). However, CNEs 1 to 3 are not equally accessible. In ATAC-seq profiles, CNEs 2 and 3 are open, consistent with data from developing mouse (95–97) and human (98) retinas, but CNE1 has a closed configuration. It may thus function as a repressive element to inhibit Atoh7 transcription in proliferating RPCs (99), mature retinal neurons, or nonretinal tissues, or it may have an architectural role (94); however, CNE1 is unlikely to regulate neighboring genes as these are separated from the Atoh7 synteny block in teleosts (26). Indeed, CNE1 has sequence similarity with a repressive element upstream of Xenopus homolog Ath5, which prevents precocious retinal expression (70). In addition, one open chromatin peak in the mouse ATAC-seq profile, 0.7 kb upstream from Di, is not conserved at the DNA sequence level (100) and is not found in human embryonic retinal open chromatin (98). If this element augments activity of the mouse Atoh7 PE, it may help explain the species difference in SE deletion phenotypes.
One function of dual Atoh7 enhancers may be to modulate chromatin looping and accessibility over time as progenitors progress through a series of competence states (101). Remarkably, the SE deletion did not alter the E14.5 retinal ATAC-seq profiles of chromatin surrounding Atoh7 or other biologically relevant genes. The SE thus does not control or gate access of transcription factors to the PE or promoter. Indeed, if these two enhancers acted cooperatively and looped to form a single holo complex with transcription factors at the Atoh7 promoter, the SE deletion would be expected to reduce accessibility of PE chromatin, as has been observed for hypersensitive sites in the β-globin locus control region (LCR) (102). Instead, the Atoh7 SEs and PEs appear to act redundantly to rapidly boost expression, similar to mouse rod opsin enhancers (40), β-globin and cone opsin LCRs (103–105), and dual Pax6 lens enhancers (44, 104). In this respect, the Atoh7 SE functionally resembles SEs originally defined in Drosophila for kruppel, knirps, and svb genes (36, 37, 106). This tandem arrangement helps establish a robust RGC progenitor pool for the developing eye.
Methods
Animals.
Atoh7SE mutant mice were generated by injecting fertilized C57BL/6J oocytes with CRISPR/Cas9 ribonucleoprotein (RNP) complexes that cleave genomic sites flanking the Atoh7 SE (SI Appendix, Supplemental Methods and Table S6). Two further alleles were combined with Atoh7SE on an isogenic C57 background in different experiments. Atoh7lacZ (Atoh7tm1Gla) is a null (KO) allele with in-frame lacZ reporter (16). Atoh7HA has three tandem HA epitope tags at the protein C terminus, intact 3′ untranslated region (UTR), and WT activity (45).
Triplex RT-PCR.
Total RNA from HA/+ and SE/+ littermate retinas (E11.5, E14.5, E16.5, and P0.5) was used to direct complementary DNA (cDNA) synthesis (SI Appendix, Supplemental Methods), and PCR was performed using three primers—one shared forward primer end-labeled with 6-carboxyfluorescein (FAM) and two reverse primers specific for UT (WT, SE) or HA alleles (SI Appendix, Table S6). Denatured products were resolved by capillary electrophoresis. Peak areas reflect the molar abundance of UT and HA cDNAs. Since Atoh7 has no introns, we included an HA/+ genomic DNA (gDNA) control to assess differential amplification of UT and HA alleles.
Immunostaining, Embryonic Cell Counts, EdU Labeling, and Optic Nerve Histology.
Tissue cryosections (10 μm) were prepared and stained, following standard procedures (SI Appendix, Supplemental Methods and Table S7), and imaged using a Leica SPE confocal microscope (Wetzlar, Germany). Optical sections were collected every 2 µm, producing a layered Z-stack. Composite Atoh7, Isl1/2, Brn3, Rbpms, c-PARP, and Thrβ2 images were stitched from nasal and temporal retinal fields using Leica LASX software. Cells were counted in maximum intensity projections of two midlevel optical planes, with n ≥ 3 biological replicates and equal exposures for all genotypes. Planimetric densities were determined using neuroretinal areas, calculated by Fiji (107). To evaluate the onset of Atoh7 expression during terminal S-phase, E14.5 gravid females were injected 90 min before harvest with 12 μg/g EdU, which was detected in fixed nuclei using Alexa Fluor 594 azide.
Ultrathin optic nerve sections from P30 and P1 mice (n = 3 +/+, SE/SE, SE/KO) were stained with toluidine blue or processed for electron microscopy to determine cross-section areas and myelinated axon counts (SI Appendix, Supplemental Methods) (19). P30 and P1 optic nerve areas were calculated by perimeter tracing (Fiji). P30 myelinated axons were counted (19, 108) for each nerve at 14 locations (10 peripheral, four central). Counts per field were averaged for each sample, and axons per nerve were calculated as mean axon density × area.
Retinal Flatmounts.
Adult retinas were dissected, fixed and flattened (photoreceptors downward), and immunostained (SI Appendix, Supplemental Methods and Table S7). After staining, retinas were mounted (RGCs facing up) and imaged at 200× using an ApoTome, collecting six individual planes in 2-µm steps. Images were tiled to form whole-retina composites and Z-stacks compressed as maximum intensity projections (ZenPro; Zeiss). To count and map RGCs, retinal flatmounts were stained with anti-Rbpms. Composite images were traced to generate area masks (Fiji) and processed to enumerate positive cells and plot RGC density using custom Python codes (SI Appendix, Supplemental Methods). Automated and manual counts were highly correlated (SI Appendix, Fig. S6). In flatmount images, orientation is indicated with D (dorsal), V (ventral), N (nasal), and T (temporal) axes.
Live OCT.
Using custom OCT system hardware and methods (109, 110), we imaged fundi of anesthetized mice in phase variance mode (pv-OCT), capturing multiple consecutive B-scans per retina position to visualize retinal blood flow and map vasculature. Scans were processed extensively to generate sectional intensity and phase variance images and depth-coded en face views (SI Appendix, Supplemental Methods). Images were processed using Matlab (Mathworks, Natick, MA) and Fiji software. Retinal B-scans were processed, to generate intensity and phase variance images, and flattened using a custom Matlab script. Intensity images were cropped to include the NFL and resliced to generate en face views. pv-OCT images were cropped to encompass the inner retina and coded for depth.
ATAC-Seq.
Dissected pairs of E14.5 +/+ and SE/SE retinas (n = 3 each) were processed into single cell suspensions (SI Appendix, Supplemental Methods). For each sample, 100,000 cells were exposed to Tn5 transposase and extracted following a scaled ATAC-seq protocol (68) with the Nextera DNA Library Prep Kit (Illumina, San Diego, CA). ATAC-seq libraries were amplified from transposed genomic DNA using barcoded primers (SI Appendix, Table S6) and established guidelines (SI Appendix, Supplemental Methods), with quality assessment by Qubit assay, high sensitivity DNA bioanalyzer (Agilent, Santa Clara, CA), and qPCR (KAPA Library Quantification; Roche, Indianapolis, IN). The six sample libraries were pooled (300 nM each) and size-selected (1.8× ratio SPRI beads; Beckman Coulter, Brea, CA), giving a final library with 1.9 ng/µL (17.5 nM) DNA and 300-base pair (bp) mean fragment size. This was sequenced using an Illumina HiSeq4000 instrument with paired-end 100 format (Novogene, Sacramento, CA) and analyzed to generate bigWig alignment files (SI Appendix, Supplemental Methods).
Statistics.
Genotype groups were compared using Student’s t test and Tukey multiple comparison tests, assuming cell count and mRNA data are normally distributed, with nonparametric bootstrap confirmation (pooled resampling × 1,000 iterations) implemented in R (111–113). Error is reported as SD. Parametric significance is indicated in display figures as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Supplementary Material
Acknowledgments
We thank Lina Du (Harvard Medical School Transgenic Core) for performing pronuclear injections to generate Atoh7 SE deletion mice, Xiuqian Mu for Atoh7HA knock-in mice, Jie Li for bioinformatics help with ATAC-seq analysis, Brad Shibata for help with EM processing and imaging, Marie-Audrey Kautzmann for early contributions to the study, and Pradhan Hariharan for counting axons. This work was supported by NIH R01 Grants EY019497 (to T.G.), EY13612 (to N.L.B.), and EY029087 (to N.M.-A.); NIH National Research Service Award F32 EY028003 and National Eye Institute Training Grant T32EY015387 (to J.B.M.); and NIH Core Facilities Grant P30EY012576.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.C.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006888117/-/DCSupplemental.
Data Availability.
The automated cell counting (cellcounterv9.py) and density heatmap (cellheatmap_v3BF.py) computer codes are available at https://github.com/jbmiesfeld/Atoh7-remote-enhancer. ATAC-seq data were deposited at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (accession no. GSE146897).
References
- 1.La Vail M. M., Rapaport D. H., Rakic P., Cytogenesis in the monkey retina. J. Comp. Neurol. 309, 86–114 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Turner D. L., Cepko C. L., A common progenitor for neurons and glia persists in rat retina late in development. Nature 328, 131–136 (1987). [DOI] [PubMed] [Google Scholar]
- 3.Wang Y., Dakubo G. D., Thurig S., Mazerolle C. J., Wallace V. A., Retinal ganglion cell-derived sonic hedgehog locally controls proliferation and the timing of RGC development in the embryonic mouse retina. Development 132, 5103–5113 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Dakubo G. D. et al., Retinal ganglion cell-derived sonic hedgehog signaling is required for optic disc and stalk neuroepithelial cell development. Development 130, 2967–2980 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Edwards M. M. et al., The deletion of Math5 disrupts retinal blood vessel and glial development in mice. Exp. Eye Res. 96, 147–156 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Sullivan M. L. et al., Astrocytes follow ganglion cell axons to establish an angiogenic template during retinal development. Glia 65, 1697–1716 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang B., Bezhadian M. A., Caldwell R. B., Astrocytes modulate retinal vasculogenesis: Effects on endothelial cell differentiation. Glia 15, 1–10 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Brzezinski J. A. 4th, Prasov L., Glaser T., Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev. Biol. 365, 395–413 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanekar S. et al., Xath5 participates in a network of bHLH genes in the developing Xenopus retina. Neuron 19, 981–994 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Brown N. L. et al., Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development 125, 4821–4833 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Kay J. N., Link B. A., Baier H., Staggered cell-intrinsic timing of ath5 expression underlies the wave of ganglion cell neurogenesis in the zebrafish retina. Development 132, 2573–2585 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Le T. T., Wroblewski E., Patel S., Riesenberg A. N., Brown N. L., Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev. Biol. 295, 764–778 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Kiyama T. et al., Overlapping spatiotemporal patterns of regulatory gene expression are required for neuronal progenitors to specify retinal ganglion cell fate. Vision Res. 51, 251–259 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miesfeld J. B., Glaser T., Brown N. L., The dynamics of native Atoh7 protein expression during mouse retinal histogenesis, revealed with a new antibody. Gene Expr. Patterns 27, 114–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kay J. N., Finger-Baier K. C., Roeser T., Staub W., Baier H., Retinal ganglion cell genesis requires lakritz, a Zebrafish atonal Homolog. Neuron 30, 725–736 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Brown N. L., Patel S., Brzezinski J., Glaser T., Math5 is required for retinal ganglion cell and optic nerve formation. Development 128, 2497–2508 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S. W. et al., Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 15, 24–29 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasov L. et al., ATOH7 mutations cause autosomal recessive persistent hyperplasia of the primary vitreous. Hum. Mol. Genet. 21, 3681–3694 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prasov L., Nagy M., Rudolph D. D., Glaser T., Math5 (Atoh7) gene dosage limits retinal ganglion cell genesis. Neuroreport 23, 631–634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng L. et al., MATH5 controls the acquisition of multiple retinal cell fates. Mol. Brain 3, 36 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riesenberg A. N. et al., Pax6 regulation of Math5 during mouse retinal neurogenesis. Genesis 47, 175–187 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miesfeld J. B. et al., Rbpj direct regulation of Atoh7 transcription in the embryonic mouse retina. Sci. Rep. 8, 10195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skowronska-Krawczyk D., Ballivet M., Dynlacht B. D., Matter J. M., Highly specific interactions between bHLH transcription factors and chromatin during retina development. Development 131, 4447–4454 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Del Bene F. et al., In vivo validation of a computationally predicted conserved Ath5 target gene set. PLoS Genet. 3, 1661–1671 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutcheson D. A. et al., bHLH-dependent and -independent modes of Ath5 gene regulation during retinal development. Development 132, 829–839 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Ghiasvand N. M. et al., Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat. Neurosci. 14, 578–586 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keser V. et al., The genetic causes of nonsyndromic congenital retinal detachment: A genetic and phenotypic study of Pakistani families. Invest. Ophthalmol. Vis. Sci. 58, 1028–1036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan K. et al., Next generation sequencing identifies mutations in Atonal homolog 7 (ATOH7) in families with global eye developmental defects. Hum. Mol. Genet. 21, 776–783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atac D. et al., Atonal homolog 7 (ATOH7) loss-of-function mutations in predominant bilateral optic nerve hypoplasia. Hum. Mol. Genet. 29, 132–148 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Brzezinski J. A. 4th et al., Loss of circadian photoentrainment and abnormal retinal electrophysiology in Math5 mutant mice. Invest. Ophthalmol. Vis. Sci. 46, 2540–2551 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macgregor S. et al., Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum. Mol. Genet. 19, 2716–2724 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nannini D. R. et al., A genome-wide association study of vertical cup-disc ratio in a latino population. Invest. Ophthalmol. Vis. Sci. 58, 87–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramdas W. D. et al., A genome-wide association study of optic disc parameters. PLoS Genet. 6, e1000978 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khor C. C. et al., Genome-wide association studies in Asians confirm the involvement of ATOH7 and TGFBR3, and further identify CARD10 as a novel locus influencing optic disc area. Hum. Mol. Genet. 20, 1864–1872 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Springelkamp H. et al.; Blue Mountains Eye Study—GWAS group; NEIGHBORHOOD Consortium; Wellcome Trust Case Control Consortium 2 (WTCCC2) , Meta-analysis of genome-wide association studies identifies novel loci that influence cupping and the glaucomatous process. Nat. Commun. 5, 4883 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong J. W., Hendrix D. A., Levine M. S., Shadow enhancers as a source of evolutionary novelty. Science 321, 1314 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frankel N. et al., Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490–493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lagha M., Bothma J. P., Levine M., Mechanisms of transcriptional precision in animal development. Trends Genet. 28, 409–416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perry M. W., Boettiger A. N., Bothma J. P., Levine M., Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 20, 1562–1567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corbo J. C. et al., CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 20, 1512–1525 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barolo S., Shadow enhancers: Frequently asked questions about distributed cis-regulatory information and enhancer redundancy. BioEssays 34, 135–141 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long H. K., Prescott S. L., Wysocka J., Ever-changing landscapes: Transcriptional enhancers in development and evolution. Cell 167, 1170–1187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong N., Kang C., Raulet D. H., Redundant and unique roles of two enhancer elements in the TCRgamma locus in gene regulation and gammadelta T cell development. Immunity 16, 453–463 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Antosova B. et al., The gene regulatory network of lens induction is wired through Meis-dependent shadow enhancers of Pax6. PLoS Genet. 12, e1006441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu X. et al., Epitope-tagging Math5 and Pou4f2: New tools to study retinal ganglion cell development in the mouse. Dev. Dyn. 238, 2309–2317 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baye L. M., Link B. A., Nuclear migration during retinal development. Brain Res. 1192, 29–36 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elshatory Y., Deng M., Xie X., Gan L., Expression of the LIM-homeodomain protein Isl1 in the developing and mature mouse retina. J. Comp. Neurol. 503, 182–197 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu F. et al., Two transcription factors, Pou4f2 and Isl1, are sufficient to specify the retinal ganglion cell fate. Proc. Natl. Acad. Sci. U.S.A. 112, E1559–E1568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan L., Deng M., Xie X., Gan L., ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development 135, 1981–1990 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mu X., Fu X., Beremand P. D., Thomas T. L., Klein W. H., Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc. Natl. Acad. Sci. U.S.A. 105, 6942–6947 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prasov L., Glaser T., Dynamic expression of ganglion cell markers in retinal progenitors during the terminal cell cycle. Mol. Cell. Neurosci. 50, 160–168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alexiades M. R., Cepko C., Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev. Dyn. 205, 293–307 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Sinitsina V. F., [DNA synthesis and cell population kinetics in embryonal histogenesis of the retina in mice]. Arkh. Anat. Gistol. Embriol. 61, 58–67 (1971). Russian. [PubMed] [Google Scholar]
- 54.Young R. W., Cell proliferation during postnatal development of the retina in the mouse. Brain Res. 353, 229–239 (1985). [DOI] [PubMed] [Google Scholar]
- 55.O’Leary D. D. M., Fawcett J. W., Cowan W. M., Topographic targeting errors in the retinocollicular projection and their elimination by selective ganglion cell death. J. Neurosci. 6, 3692–3705 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Erkman L. et al., A POU domain transcription factor-dependent program regulates axon pathfinding in the vertebrate visual system. Neuron 28, 779–792 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Dangata Y. Y., Findlater G. S., Kaufman M. H., Postnatal development of the optic nerve in (C57BL x CBA)F1 hybrid mice: General changes in morphometric parameters. J. Anat. 189, 117–125 (1996). [PMC free article] [PubMed] [Google Scholar]
- 58.Rodriguez A. R., de Sevilla Müller L. P., Brecha N. C., The RNA binding protein RBPMS is a selective marker of ganglion cells in the mammalian retina. J. Comp. Neurol. 522, 1411–1443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zarei K., et al. , Automated axon counting in rodent optic nerve sections with AxonJ. Sci. Rep. 6, 26559 (2016). Correction in: Sci. Rep. 6, 34124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salinas-Navarro M. et al., Retinal ganglion cell population in adult albino and pigmented mice: A computerized analysis of the entire population and its spatial distribution. Vision Res. 49, 637–647 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Dräger U. C., Olsen J. F., Ganglion cell distribution in the retina of the mouse. Invest. Ophthalmol. Vis. Sci. 20, 285–293 (1981). [PubMed] [Google Scholar]
- 62.Salinas-Navarro M. et al., A computerized analysis of the entire retinal ganglion cell population and its spatial distribution in adult rats. Vision Res. 49, 115–126 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Morcillo J. et al., Proper patterning of the optic fissure requires the sequential activity of BMP7 and SHH. Development 133, 3179–3190 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Tao C., Zhang X., Development of astrocytes in the vertebrate eye. Dev. Dyn. 243, 1501–1510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mi H., Barres B. A., Purification and characterization of astrocyte precursor cells in the developing rat optic nerve. J. Neurosci. 19, 1049–1061 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burne J. F., Raff M. C., Retinal ganglion cell axons drive the proliferation of astrocytes in the developing rodent optic nerve. Neuron 18, 223–230 (1997). [DOI] [PubMed] [Google Scholar]
- 67.Hellström A., Wiklund L. M., Svensson E., Albertsson-Wikland K., Strömland K., Optic nerve hypoplasia with isolated tortuosity of the retinal veins: A marker of endocrinopathy. Arch. Ophthalmol. 117, 880–884 (1999). [DOI] [PubMed] [Google Scholar]
- 68.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1-21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willardsen M., Hutcheson D. A., Moore K. B., Vetter M. L., The ETS transcription factor Etv1 mediates FGF signaling to initiate proneural gene expression during Xenopus laevis retinal development. Mech. Dev. 131, 57–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Willardsen M. I. et al., Temporal regulation of Ath5 gene expression during eye development. Dev. Biol. 326, 471–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hay D. et al., Genetic dissection of the α-globin super-enhancer in vivo. Nat. Genet. 48, 895–903 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao Z., Mao C. A., Pan P., Mu X., Klein W. H., Transcriptome of Atoh7 retinal progenitor cells identifies new Atoh7-dependent regulatory genes for retinal ganglion cell formation. Dev. Neurobiol. 74, 1123–1140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Provis J. M., van Driel D., Billson F. A., Russell P., Human fetal optic nerve: Overproduction and elimination of retinal axons during development. J. Comp. Neurol. 238, 92–100 (1985). [DOI] [PubMed] [Google Scholar]
- 74.Rakic P., Riley K. P., Overproduction and elimination of retinal axons in the fetal rhesus monkey. Science 219, 1441–1444 (1983). [DOI] [PubMed] [Google Scholar]
- 75.Strom R. C., Williams R. W., Cell production and cell death in the generation of variation in neuron number. J. Neurosci. 18, 9948–9953 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farah M. H., Easter S. S. Jr., Cell birth and death in the mouse retinal ganglion cell layer. J. Comp. Neurol. 489, 120–134 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Brzezinski J. A. et al., Math5 null mice have abnormal retinal and persistent hyaloid vasculatures. Dev. Biol. 259, 394 (2003). [Google Scholar]
- 78.Campbell I. C., Coudrillier B., Ross Ethier C., Biomechanics of the posterior eye: A critical role in health and disease. J. Biomech. Eng. 136, 021005 (2014). [DOI] [PubMed] [Google Scholar]
- 79.David T., Smye S., James T., Dabbs T., Time-dependent stress and displacement of the eye wall tissue of the human eye. Med. Eng. Phys. 19, 131–139 (1997). [DOI] [PubMed] [Google Scholar]
- 80.Liu W., Mo Z., Xiang M., The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc. Natl. Acad. Sci. U.S.A. 98, 1649–1654 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hutcheson D. A., Vetter M. L., The bHLH factors Xath5 and XNeuroD can upregulate the expression of XBrn3d, a POU-homeodomain transcription factor. Dev. Biol. 232, 327–338 (2001). [DOI] [PubMed] [Google Scholar]
- 82.Prasov L., Glaser T., Pushing the envelope of retinal ganglion cell genesis: Context dependent function of Math5 (Atoh7). Dev. Biol. 368, 214–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang T., Ranade S., Cai C. Q., Clouser C., Pignoni F., Direct control of neurogenesis by selector factors in the fly eye: Regulation of atonal by Ey and so. Development 133, 4881–4889 (2006). [DOI] [PubMed] [Google Scholar]
- 84.Sekido R., Lovell-Badge R., Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453, 930–934 (2008). [DOI] [PubMed] [Google Scholar]
- 85.Bullejos M., Koopman P., Delayed Sry and Sox9 expression in developing mouse gonads underlies B6-Y(DOM) sex reversal. Dev. Biol. 278, 473–481 (2005). [DOI] [PubMed] [Google Scholar]
- 86.Hiramatsu R. et al., A critical time window of Sry action in gonadal sex determination in mice. Development 136, 129–138 (2009). [DOI] [PubMed] [Google Scholar]
- 87.Larney C., Bailey T. L., Koopman P., Switching on sex: Transcriptional regulation of the testis-determining gene Sry. Development 141, 2195–2205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim J. et al., GDF11 controls the timing of progenitor cell competence in developing retina. Science 308, 1927–1930 (2005). [DOI] [PubMed] [Google Scholar]
- 89.La Torre A., Georgi S., Reh T. A., Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, E2362–E2370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Riesenberg A. N., Liu Z., Kopan R., Brown N. L., Rbpj cell autonomous regulation of retinal ganglion cell and cone photoreceptor fates in the mouse retina. J. Neurosci. 29, 12865–12877 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wiencken-Barger A. E., Mavity-Hudson J., Bartsch U., Schachner M., Casagrande V. A., The role of L1 in axon pathfinding and fasciculation. Cereb. Cortex 14, 121–131 (2004). [DOI] [PubMed] [Google Scholar]
- 92.Jiang Y. et al., Transcription factors SOX4 and SOX11 function redundantly to regulate the development of mouse retinal ganglion cells. J. Biol. Chem. 288, 18429–18438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nord A. S., West A. E., Neurobiological functions of transcriptional enhancers. Nat. Neurosci. 23, 5–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Klemm S. L., Shipony Z., Greenleaf W. J., Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 95.Aldiri I. et al., The dynamic epigenetic landscape of the retina during development, reprogramming, and tumorigenesis. Neuron 94, 550–568.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wilken M. S. et al., DNase I hypersensitivity analysis of the mouse brain and retina identifies region-specific regulatory elements. Epigenetics Chromatin 8, 8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zibetti C., Liu S., Wan J., Qian J., Blackshaw S., Epigenomic profiling of retinal progenitors reveals LHX2 is required for developmental regulation of open chromatin. Commun. Biol. 2, 142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cherry T. J. et al., Mapping the cis-regulatory architecture of the human retina reveals noncoding genetic variation in disease. Proc. Natl. Acad. Sci. U.S.A. 117, 9001–9012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sinn R., Peravali R., Heermann S., Wittbrodt J., Differential responsiveness of distinct retinal domains to Atoh7. Mech. Dev. 133, 218–229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Villar D. et al., Enhancer evolution across 20 mammalian species. Cell 160, 554–566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tolhuis B., Palstra R. J., Splinter E., Grosveld F., de Laat W., Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10, 1453–1465 (2002). [DOI] [PubMed] [Google Scholar]
- 102.Forrester W. C. et al., A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 4, 1637–1649 (1990). [DOI] [PubMed] [Google Scholar]
- 103.Cimbora D. M. et al., Long-distance control of origin choice and replication timing in the human beta-globin locus are independent of the locus control region. Mol. Cell. Biol. 20, 5581–5591 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bender M. A., Bulger M., Close J., Groudine M., β-globin gene switching and DNase I sensitivity of the endogenous β-globin locus in mice do not require the locus control region. Mol. Cell 5, 387–393 (2000). [DOI] [PubMed] [Google Scholar]
- 105.Wang Y. et al., Mutually exclusive expression of human red and green visual pigment-reporter transgenes occurs at high frequency in murine cone photoreceptors. Proc. Natl. Acad. Sci. U.S.A. 96, 5251–5256 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.El-Sherif E., Levine M., Shadow enhancers mediate dynamic shifts of gap gene expression in the Drosophila embryo. Curr. Biol. 26, 1164–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schindelin J. et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Williams R. W., Strom R. C., Rice D. S., Goldowitz D., Genetic and environmental control of variation in retinal ganglion cell number in mice. J. Neurosci. 16, 7193–7205 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim D. Y. et al., In vivo volumetric imaging of human retinal circulation with phase-variance optical coherence tomography. Biomed. Opt. Express 2, 1504–1513 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang P. et al., In vivo wide-field multispectral scanning laser ophthalmoscopy-optical coherence tomography mouse retinal imager: Longitudinal imaging of ganglion cells, microglia, and Müller glia, and mapping of the mouse retinal and choroidal vasculature. J. Biomed. Opt. 20, 126005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.R Core Team , R: A Language and Environment for Statistical Computing, (Version 3.5.1, R Foundation for Statistical Computing, Vienna, 2013). [Google Scholar]
- 112.Dwivedi A. K., Mallawaarachchi I., Alvarado L. A., Analysis of small sample size studies using nonparametric bootstrap test with pooled resampling method. Stat. Med. 36, 2187–2205 (2017). [DOI] [PubMed] [Google Scholar]
- 113.Tibshirani B. E. R. J., An Introduction to the Bootstrap (Monographs on Statistics and Applied Probability, Chapman & Hall CRC, Boca Raton, FL, 1993), vol. 57. [Google Scholar]
- 114.Ovcharenko I. et al., ECR browser: A tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 32, W280–W286 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The automated cell counting (cellcounterv9.py) and density heatmap (cellheatmap_v3BF.py) computer codes are available at https://github.com/jbmiesfeld/Atoh7-remote-enhancer. ATAC-seq data were deposited at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (accession no. GSE146897).