Fig. 1.
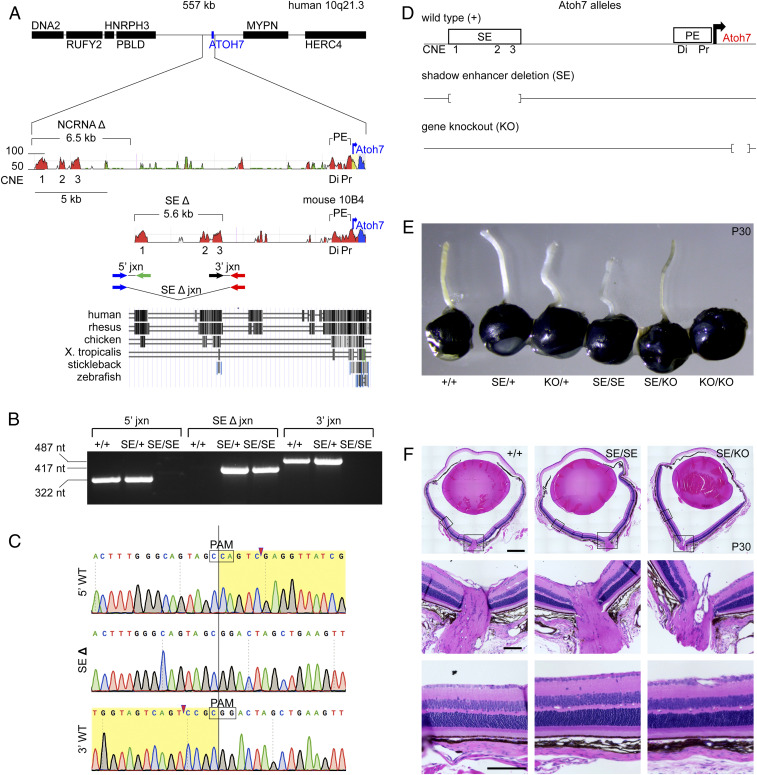
Targeted deletion of the murine Atoh7 SE. (A) Human and mouse Atoh7 locus maps. (Top) VISTA plots (114) show conserved noncoding elements within SEs (CNE1-3) and PEs (Di, Pr). Human NCRNA and mouse SE deletions (brackets) and transcription start sites (blue arrows) are indicated. Sequence identity (50 to 100%) is plotted for a 50-bp moving window. DNA segments are identified as intergenic (red), repetitive (green), coding (blue), or UTR (yellow). Primers used to amplify SE deletion (Δ) junction and WT endpoints are indicated below the mouse VISTA plot. (Bottom) Multiz species alignment, adapted from the University of California Santa Cruz (UCSC) browser (build mm10). (B) Agarose gel showing diagnostic PCRs from +/+, SE/+, and SE/SE genomic DNA. (C) Sanger chromatograms of PCR products spanning the SEΔ junction and WT endpoints. Deleted nucleotides (yellow highlight), Cas9 cut sites (arrowheads), and protospacer adjacent motifs (PAMs) are indicated. (D) Schema of Atoh7 alleles. (E) Adult eyes from Atoh7 genotypic series. The optic nerves are thin in SE/KO eyes (with 56.6 ± 10.6% of +/+ cross-sectional area, SI Appendix, Table S4) and absent in KO/KO eyes. (F) Histology of adult eyes (hematoxylin/eosin) with similar retinal morphology. X. tropicalis, Xenopus tropicalis; nt, nucleotides. (Scale bars: Top, 500 µm; and Middle and Bottom, 100 µm.)