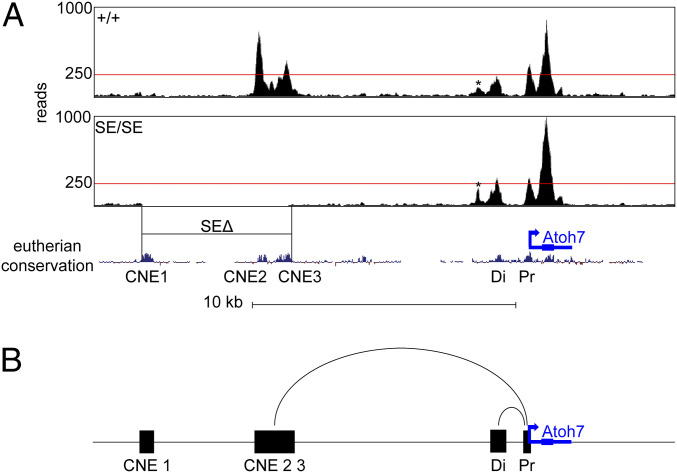
Fig. 7.
Open chromatin signatures surrounding Atoh7. (A) (A) ATAC-seq profiles of +/+ and SE/SE mutant E14.5 retinas across the Atoh7 locus. The total number of aligned reads per nucleotide are shown as bigWig tracks in the UCSC mm10 genome browser, with 250 reads indicated (red line). Each plot reflects pooled data from three embryo replicates (six retinas, >69 million reads). Open chromatin peaks are apparent over CNE2, CNE3, Di, Pr, a small nonconserved element (*) and the Atoh7gene body. No reads were mapped to CNE2 or CNE3 in SE/SE chromatin; otherwise, the ATAC-seq profiles are similar. The open chromatin status and accessibility of PE does not depend on SE. (B) Proposed mechanism for Atoh7 regulation by SE and PE during retinal histogenesis. SE (CNE2/3 elements) and PE (distal element) loop independently with the Atoh7 promoter to activate transcription and are functionally redundant. Each modular enhancer is sufficient to control spatiotemporal expression, but they act additively to increase activity.