Fig. 4.
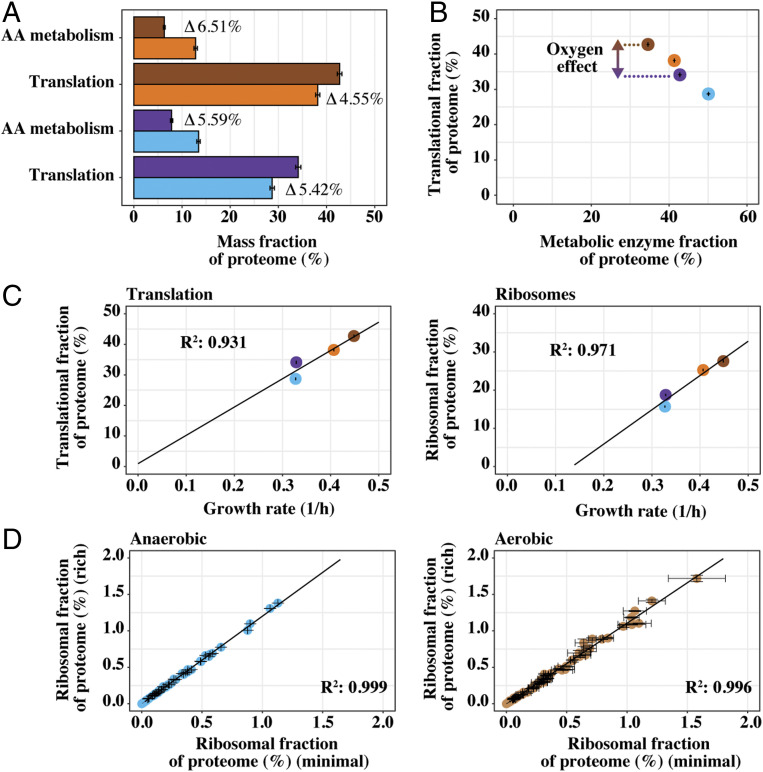
The decreased proteome allocation for amino acid biosynthesis is partly directed toward protein translation. (A) Average summed allocation, with SD, of yeast proteins in translation and amino acid (AA) metabolism, respectively. Differences in allocation, between rich and minimal medium, are highlighted for both anaerobic (n = 3) and aerobic (n = 2) conditions. (B) Average mass allocation, with SD, of translation-related proteins versus all quantified enzymes related to metabolism. (C) Summed allocation of translation and ribosomal proteins, with SD, versus growth rate (n = 3 for anaerobic and n = 2 for aerobic conditions). (D) Comparison of mean allocation, with SD, for all individual ribosomal proteins between cells cultivated in rich medium versus minimal medium (n = 3 for anaerobic and n = 2 for aerobic conditions). Proteins assigned to each group (translation, AA metabolism, ribosomes, and metabolic enzymes) were based on subsetting as performed in ref. 26. Metabolic enzymes reflected groups: AA_metabolism, Energy, Lipids, Nucleotides, Mitochondria and Glycolysis. Colors are as in Fig. 3E.