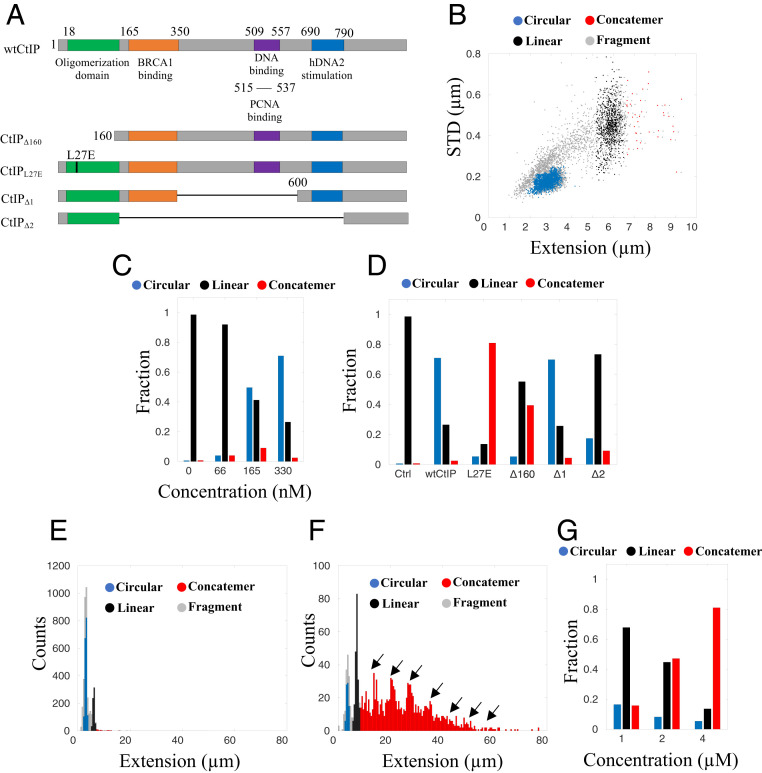
Fig. 2.
The tetrameric structure of CtIP is important for circularization of λ-DNA. (A) Schematic illustration of the CtIP derivatives used in this study, with color-coded regions highlighting different domains of interest. The numbers correspond to the position in the protein sequence. (B) Scatterplot of molecule extension vs. STD for λ-DNA (4 µM bp) incubated with wtCtIP (330 nM), equivalent to 500 tetramers per DNA end (n = 3,886). Clustering of the datasets was performed to distinguish the circularized λ-DNA molecules (blue) from the full-size linear λ-DNA molecules (black), concatemers (red), and linear fragments (gray). (C and D) Relative fractions of circular and linear complexes and concatemers at a DNA concentration of 4 µM bp for different concentrations of wtCtIP (C) and different derivatives of the protein at a constant protein concentration of 330 nM (D). (E and F) Size histograms for wtCtIP (n = 3,886; bin size = 0.5 µm) (E) and CtIPL27E (n = 1,541; bin size = 0.5 µm) (F) at 330 nM protein and 4 µM bp DNA (concentration equivalent to 500 tetramers or 1,000 dimers per DNA end, respectively). The high frequency of concatemers for CtIPL27E compared with wtCtIP is denoted by arrows in the histogram. (G) Relative fractions of circular and linear complexes and concatemers at different concentrations of λ-DNA in the presence of CtIPL27E at a constant DNA:protein ratio corresponding to 1,000 dimers per DNA end. Increasing the DNA concentration promotes concatemer formation.