Fig. 7.
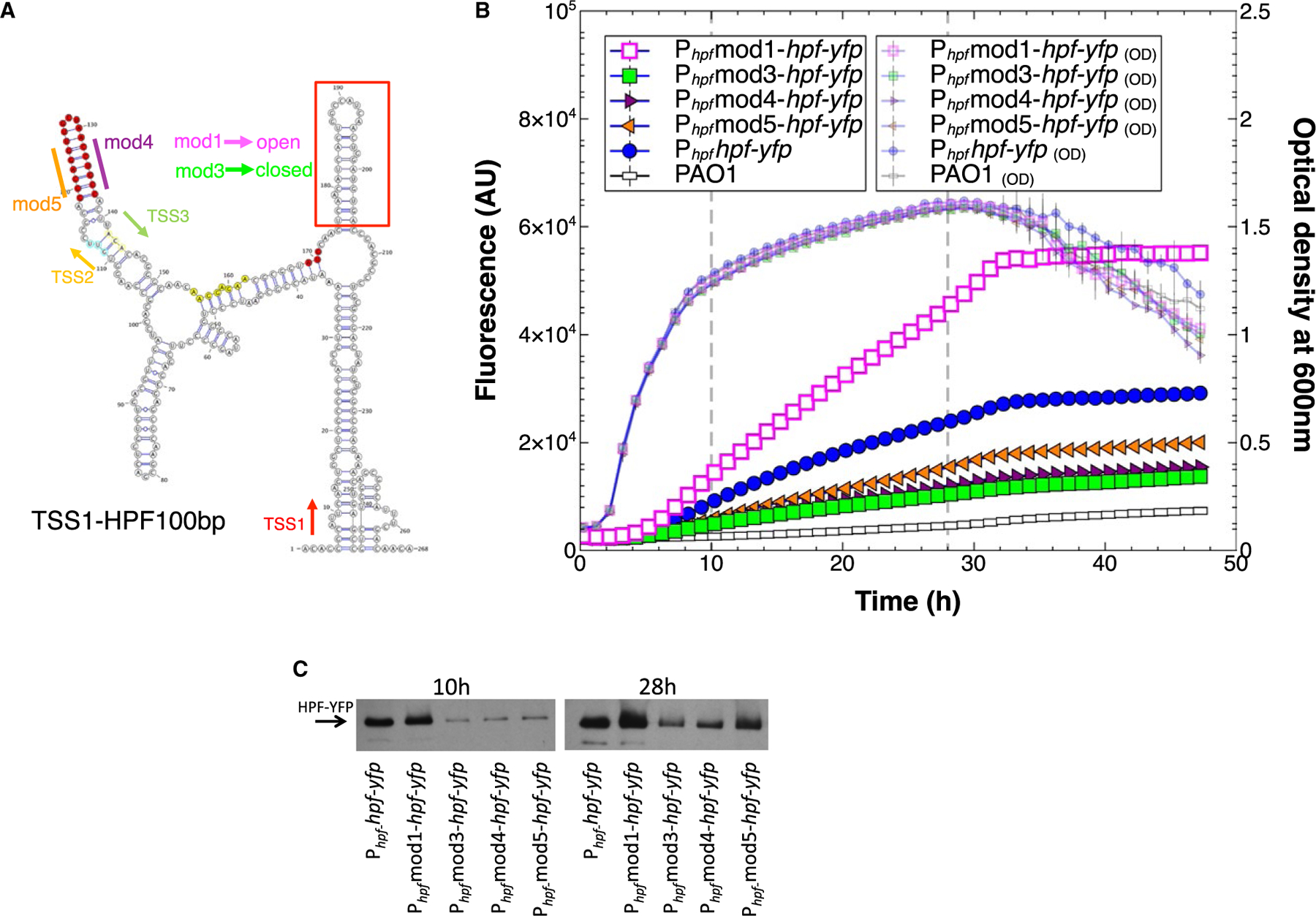
A. Secondary structure prediction of the hpf mRNA starting from TSS1 through the first 100 nucleotides of the hpf structural gene, predicted by RNAfold server and visualized using VARNA (Lorenz et al., 2011). Shown are the sites of the predicted transcriptional start sites (TSS1,2,3), and the position of the hpf start codon (red). The hairpin loop (HPL) at 5′ end of the hpf coding sequence (red box) was mutated by nucleotide substitutions without modifying the amino acid sequence of HPF, creating open (mod1) or closed (mod3) HPLs. The 5ʹ and 3′ ends of the HPL stem located between TSS2 and TSS3 (highlighted in red) were altered to disrupt the HPL structure (mod 4 and mod5).
B. Growth and expression from Phpf containing modifications; mod1 (pink squares) mod3 (green squares) mod4 (maroon triangles) mod5 (orange triangles), compared to the wild-type sequence (blue circles).
C. Immunoblot analysis of the HPF-YFP fusion protein produced at 10 and 24 h of growth, when the HPLs were modified as in Fig. 7B.
