Figure 1.
The Molecular Architecture of the SARS-CoV-2 Virus
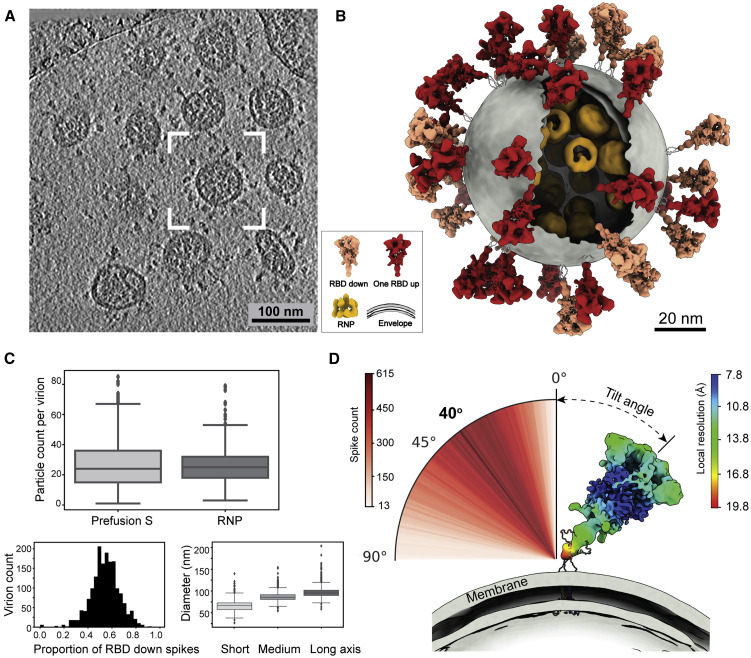
(A) A representative tomogram slice (5 Å thick) showing pleomorphic SARS-CoV-2 virions.
(B) A representative virus (boxed in A) is reconstructed by projecting the prefusion S in the “RBD down” conformation (salmon) and “one RBD up” conformation (red), the lipid envelope (gray), and RNPs (yellow) onto their refined coordinates. RNPs away from the envelope are hidden for clarity. The unsolved stem regions of the spikes are sketched from a predicted model (https://zhanglab.ccmb.med.umich.edu/COVID-19).
(C) Summary of structural observations. Top: number of prefusion Ss and RNPs per virion. An average of 26 prefusion Ss and RNPs are found in a virion. Bottom left: ratio of the S proteins between the “RBD down” and “one RBD up” conformations. An average of 54% prefusion Ss are in the “RBD down” conformation. Bottom right: statistics of the dimension of SARS-CoV-2 viral envelopes. The average diameters for the short, medium, and long axis of the envelope are 64.8, 85.9, and 96.6 nm, respectively. A boxplot shows the outliers, minimum, first quartile, medium, third quartile, and maximum of the data.
(D) Distribution of the spike tilt angle reveals a prevailing tilt of 40° relative to the normal axis of the envelope. Shown is a representative “RBD down” spike in authentic orientation to the envelope (gray). The spike is colored by local resolution, and the predicted model of the stem is fitted for illustration purpose.
See also Figure S1 and Video S1.