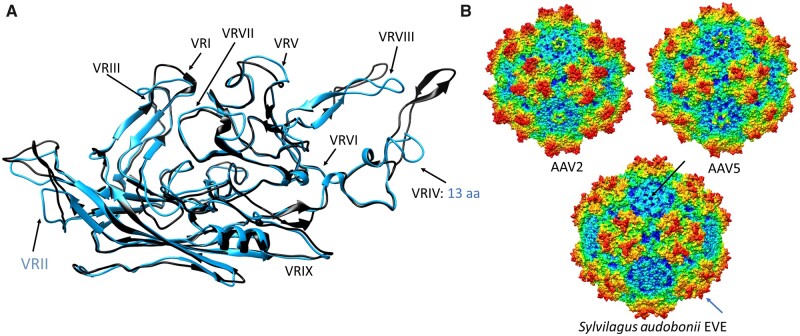
Figure 3.
Homology modelling of the capsid proteins reconstructed from the endogenous dependoparvovirus-like elements found in members of family Leporidae of order Lagomorpha, represented by the most conserved such element derived from S.audobonii. (A) Ribbon diagram of the S.audobonii capsid monomer homology model (blue) superimposed on the monomer of the AAV2 capsid (black). The blue letters indicate deletions, specifying the number of aa or the region affected, compared to the reference structure of AAV2. As the entire VRII is absent, this is highlighted in blue. (B) The complete capsid homology model of S.audobonii, with the AAV2 and 5 capsids provided for comparison. The black arrow points to the DE loop, which is missing the entire VR-II. The blue arrow highlights the truncated threefold protrusions, caused by the deletion affecting VRIV.