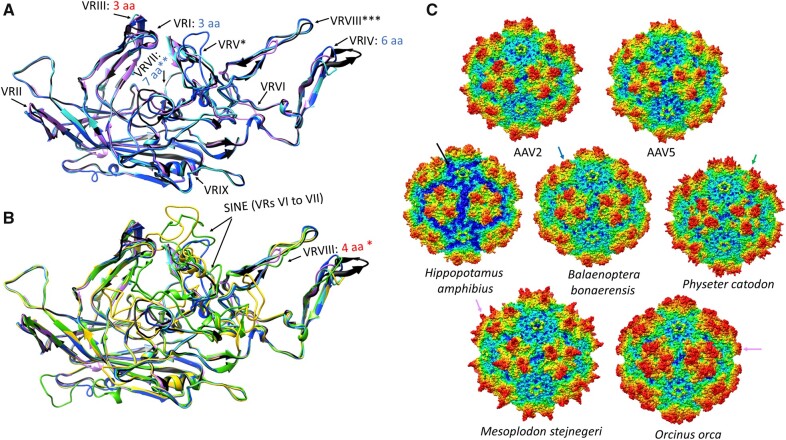
Figure 4.
Homology modelling of the capsid proteins reconstructed from the endogenous dependoparvovirus-like elements found in members of Whippomorpha. Each capsid variation is represented by one whippomorph species. VR stands for variable region. (A) Ribbon diagram superposition of the models of the most intact capsid protein monomers of the hippopotamus (pink ribbon), a baleen whale (cyan ribbon) and a basal toothed whale (blue ribbon), none of them affected by the SINE insertion. Insertions are indicated by red text, deletions by blue letters, specifying the number of aa affected, compared to AAV2, which is used as the reference structure (black ribbon). Features marked by stars are as follows: *—Mutated beyond recognition in P.catodon EVE, though present; **—Not affected in H.amphibius and B.bonaerensis; ***Distorted in H.amphibius due to the absence of the C-term. (B) Superposition of models of capsid monomers affected by the SINE insertion with those, which were not. Ribbon colors are shown as in panel A, with the addition of the O.orca (yellow ribbon) and the M.stejnegeri (green ribbon) elements. *—The four-aa-long insertion is only present in M.stejnegeri. (C) Complete capsid homology models of all the variations from panels A and B. The deleted C-terminal region of the Hippopotamus amphibious capsid model is marked by the black arrow. The blue arrow indicates the truncated threefold protrusions of the baleen whale entry, probably the closest to what could be find on the original capsid responsible for these endogenous elements. This region is heavily mutated in the basal toothwhale species, the sperm whale (P.catodon), marked by the green arrow. The magenta arrow shows how the SINE insertion distorts the threefold of the beaked whale and dolphin endogenous element capsids.