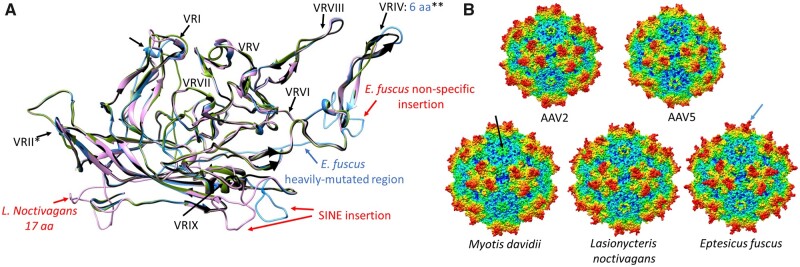
Figure 5.
Homology modelling of the capsid proteins reconstructed from the endogenous dependoparvovirus-like elements found in members of family Vespertilionidae. Each genus is represented by one entry, except for Plecotus, as this capsid protein did not significantly differ from that of any of the variations found in the other three genera. (A) Ribbon diagram superimposition of the capsid protein monomer models of the L.noctivagans (pink), Eptesicus fuscus (blue) and the M.davidii (green) elements on the AAV2 capsid monomer ribbon diagram (black). Insertions are indicated by red, deletions by blue letters, specifying the number of aa affected, compared to the reference structure of AAV2. The small hollow arrows indicate structural changes due to the missing βG sheet of the jelly roll in genus Myotis. The following features are indicated by the stars are as follows: *—A deletion of four aa in the surrounding region of genus Myotis; **—the E.fuscus element is an exception. (B) Complete capsid homology models of all the variations from section A. The black arrow points to the DE loop of M.davidii, which results in a significantly truncated fivefold channel due to the two-aa-long deletion pre- and another two within the VRII. The AAV2-like threefold peaks of E.fuscus are highlighted by the blue arrow.