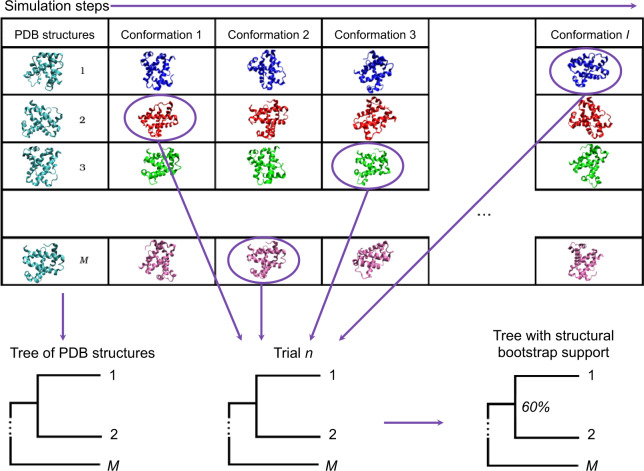
Fig. 6.
Overview of a bootstrap method for structure comparisons. An ensemble of possible conformations is generated for each of proteins using MD simulation. For each of trials, a conformation cm is randomly selected from each of the M ensembles to populate a new trial data set Cn. Pairwise comparison of the conformations in each trial data set Cn generates new distances from which a NJ tree Tn is created. Each trial tree, Tn, is compared with the reference tree T0. If a relationship between structures in the reference tree T0 is recreated in the trial tree Tn, it is counted. The nodes of T0 are labeled with the fraction of trial trees in which the relationship was recovered, providing a measure of the statistical support for that node.