Abstract
In a previous study we identified EARLY BUD BREAK 1 (EBB1), an ERF transcription factor, in peach (Prunus persica var. nectarina cultivar Zhongyou 4); however, little is known of how PpEBB1 may regulate bud break. To verify the function of PpEBB1 in bud break, PpEBB1 was transiently transformed into peach buds, resulting in early bud break. Bud break occurred earlier in PpEBB1-oe poplar (Populus trichocarpa) obtained by heterologous transformation than in wild type (WT), consistent with the peach bud results, indicating that PpEBB1 can promote bud break. To explore how PpEBB1 affects bud break, differentially expressed genes (DEGs) between WT and PpEBB1-oe poplar plants were identified by RNA-sequencing. The expression of DEGs associated with hormone metabolism, cell cycle, and cell wall modifications changed substantially according to qRT-PCR. Auxin, ABA, and total trans-zeatin-type cytokinin levels were higher in the PpEBB1-oe plants than in WT plants, while the total N6-(Δ 2-isopentenyl)-adenine-type cytokinins was lower. Yeast two-hybrid and bimolecular fluorescence complementation assays verified that a cell wall modification-related protein (PpEXBL1) interacted with PpEBB1 suggesting that PpEBB1 could interact with these cell wall modification proteins directly. Overall, our study proposed a multifaceted explanation for how PpEBB1 regulates bud break and showed that PpEBB1 promotes bud break by regulating hormone metabolism, the cell cycle, and cell wall modifications.
Keywords: Bud break, cell cycle, cell wall modification, hormone, peach (Prunus persica), PpEBB1, PpEXBL1
The transcription factor EBB1 plays important roles in regulating bud break in peach trees by regulating hormone metabolism, the cell cycle, and cell wall modifications.
Introduction
Dormancy is an important physiological phenomenon that allows trees to endure harsh environmental conditions during winter (Chen et al., 2016), during which time visible growth of any plant structure is lacking. There are three types of dormancy: paradormancy, endodormancy, and ecodormancy (Lang, 1987). In paradormancy, the growth of lateral buds is suppressed by actively growing parts of the plant, such as the apical meristem; this process is known as apical dominance. Buds in endodormancy without meeting chilling requirements will not grow outward, even under favorable conditions. Following the endodormancy stage, buds become ecodormant and resume growth when conditions become favorable (Shim et al., 2014; Castède et al., 2015). The transition from active growth to dormancy in the fall is usually induced by short-day conditions or cold temperature; the transition of peach and poplar is induced by short-day conditions. Bud dormancy is an important adaptive mechanism that can protect plants from frost damage either in late spring around the time of bud break or in early fall around the time of growth cessation (Yordanov et al., 2014).
Numerous studies have shown that plant hormones control dormancy and bud break, especially auxin, abscisic acid (ABA), gibberellin (GA), and cytokinin (Horvath et al., 2008; Balla et al., 2011; Aksenova et al., 2013; Gillespie and Volaire, 2017). Auxin is considered to play important roles in paradormancy. The classic model for the function of auxin in bud outgrowth involves the transport of auxin from the apical and lateral parts of the stem inhibiting auxin export from axillary buds, suppressing axillary bud outgrowth (Balla et al., 2011). However, there is a recent view that auxin export from axillary buds is independent of initial bud growth but is important for sustained bud outgrowth (Barbier et al., 2019). This view is also supported by research on pea (Pisum sativum) in which the application of inhibitors of both auxin transport and perception had no effects on decapitation-induced early bud outgrowth; instead, the inhibitory effects were apparent after 2 d (Chabikwa et al., 2019). ABA is recognized as the key internal factor for entering dormancy and is involved in dormancy induction and maintenance (Horvath et al., 2008; Gillespie and Volaire, 2017). The concentration of ABA decreases while that of auxin, cytokinin, and GA increases during dormancy release in herbaceous perennial species (Gillespie and Volaire, 2017). The role of GA and cytokinin application in promoting dormancy release and bud break has been widely studied (Roman et al., 2016; Gillespie and Volaire, 2017). Exogenous GA has long been used to break dormancy and promote bud burst, but the role of exogenous GA in dormancy release varies with different species (Gillespie and Volaire, 2017). Cytokinin acts as a primary regulator of dormancy release. Inactivation of cytokinin results in prolonged dormancy, while increased cytokinin activity leads to shortened dormancy (Aksenova et al., 2013). Cytokinin treatment can up-regulate the expression of GA biosynthesis genes and down-regulate the expression of GA degradation-related genes significantly (Ni et al., 2017). Therefore, crosstalk among different hormones may also play important roles in dormancy regulation. Auxin, cytokinin, and their interaction can regulate the outgrowth of axillary buds suppressed by the shoot tip (Ferguson and Beveridge, 2009). In Chrysanthemum, decreased auxin and increased cytokinin coincide with more branches, whereas high auxin levels and decreased cytokinin levels are associated with strong apical dominance (Dierck et al., 2016). In recent years, strigolactone was discovered to be a branching inhibitor, and could regulate bud outgrowth by controlling auxin export from axillary buds together with cytokinin (Crawford et al., 2010; Shinohara et al., 2013; Rameau et al., 2014; Waldie and Leyser, 2018). However, to date, the role of all these hormones during the control of growth cessation, bud dormancy, and outgrowth is still not fully understood (Maurya and Bhalerao, 2017).
When buds begin to grow outward, an increase in cell division often occurs (Horvath et al., 2003). The plant cell cycle consists of four main stages: DNA synthesis (S), mitosis (M), and two intervening gap phases (G1 and G2) (Velappan et al., 2017). Most cells in the buds are arrested at the G1 phase of the cell cycle during dormancy (Devitt and Stafstrom, 1995). Different types of cyclins (CYCs) operate in concert with cyclin-dependent kinases (CDKs) during different cell cycle stages. A-type CDKs (CDKAs) function mainly in the G1–S and G2–M phase transitions, while B-type CDKs (CDKBs) function in the G2–M phase transition. CDKAs form a complex with CYCDs during the G1–S transition, while CDKAs form a complex with CYCAs, promoting the transition to G2. CYCA/B operates together with CDKA and CDKB to regulate the G2–M transition (Velappan et al., 2017). The expression of genes related to the G1-to-S-phase transition, such as D-cyclins (CYCDs) and CDKs, is one of the conditions for bud outgrowth (Shimizu-Sato and Mori, 2001; Aksenova et al., 2013). A large number of studies have directly linked cell development to signals such as hormones that regulate adventitious bud or axillary bud dormancy (Horvath et al., 2003). There is a negative correlation between ABA content and transcript levels of cell cycle genes in grapevine latent buds and shoot apexes. The expression of cell cycle-related genes (CDKB1/2, CYCA1/2/3, CYCB, CYCD3.2a) is repressed by ABA, and the expression of cell cycle-related genes is decreased when the ABA content increases during endodormancy (Vergara et al., 2017). GA and cytokinin treatment can regulate the expression of genes related to the cell cycle, indicating that these hormones may promote bud outgrowth by regulating the cell cycle machinery in axillary buds, which was shown in Jatropha curcas (Ni et al., 2017).
Plant cell walls are important for plant growth and development, for protection against cell wall damage detection, and for bioconversion (De Lorenzo et al., 2019). Recently, studies have shown that cell walls can regulate seed germination (Bassel, 2016) and bud outgrowth in both annual and perennial plants (Lee et al., 2017). Cell walls are composed mainly of cellulose, hemicellulose, pectin, lignin, and proteins (Sarkar et al., 2009). Enzymes mediating these components, such as endotransglycosylases/hydrolases (XTHs), pectin methylesterases (PMEs), and expansins, are important for cell wall loosening and for regulating germination. In Arabidopsis seeds, the activity of cell wall enzymes plays an important role in germination, which can lead to radicle protrusion by enabling embryo cell expansion (Sechet et al., 2016). The genes related to the promotion of cell expansion are not expressed in dormant seeds (Cadman et al., 2006). In Norway spruce, the permeability of the cell wall in growing shoot tips can be affected by the photoperiod, thereby regulating bud growth–dormancy cycling by influencing the transport of water into the buds (Lee et al., 2017).
Bud dormancy has an important impact on fruit production, especially in perennial deciduous fruit trees (Sun et al., 2016). However, the key genes that function in the transition between dormancy and regrowth are still poorly understood. DORMANCY ASSOCIATED MADS-BOX (DAM) genes were initially thought to be associated with dormancy because evergrowing peach mutants, which have a deletion in a series of DAM genes, do not stop growing or develop buds under short-day conditions (Bielenberg et al., 2004, 2008). DAM genes have been identified as being critical factors controlling endodormancy and dormancy release in many species. For example, DAM5 and DAM6 in peach (Prunus persica) may function in the chilling requirement of lateral buds and may be negatively correlated with bud burst (Yamane et al., 2011). The transcript levels of DAM1 and DAM3 in pear (Pyrus pyrifolia) peak at the endodormancy stage but then decrease at the ecodormancy stage (Niu et al., 2016). A study on Prunus mume indicated that PmDAMs interact with other PmDAMs or themselves to form protein complexes (Zhao et al., 2018a). Moreover, in pear, the expression of DAM can be directly promoted by the accumulation of C-repeat binding factor (CBF) and then inhibits FLOWERING LOCUS T (FT) expression to induce endodormancy (Niu et al., 2016). Recently, EARLY BUD BREAK 1 (EBB1) was identified as promoting bud break in poplar by reactivating meristematic activity after winter dormancy (Yordanov et al., 2014), and homologs of EBB1 in apple, pear, grape, and spruce may function in bud break (Busov et al., 2016; Wisniewski et al., 2015; Anh Tuan et al., 2016). However, it remains unclear how EBB1 regulates bud break. Here, the EBB1 in peach, which was identified in our previous study based on its expression level during the dormancy phase (Zhao et al., 2018b), was heterologously transformed into poplar because there were no transformation methods in peach, and RNA-sequencing (RNA-seq) was used to investigate the mechanism by which PpEBB1 regulates bud break.
Materials and methods
Plant materials
Samples of buds of peach (Prunus persica var. nectarina) cultivar Zhongyou 4 were harvested from 5-year-old trees growing at the Horticulture Experimental Station of Shandong Agricultural University, Tai’an, China. All trees were grown in accordance with standard agricultural practices. Leaf bud samples were collected from first-year branches approximately every 2 weeks from October 2016 until April 2017. The samples were immediately flash frozen in liquid nitrogen and subsequently stored at −80 °C for total RNA extraction and qRT-PCR-based experiments.
Construction of viral vectors and transient expression in buds
Transient expression assays were performed in peach buds. The coding DNA sequence (CDS) of PpEBB1 was cloned and inserted into an IL60 virus plasmid vector. The shoots used for transient expression were randomly collected and cultivated under the following conditions: 16 h of light at 25 °C with an artificial fluorescence lamp (200 µmol m−2 s−1) and 8 h of darkness at 18 °C. The relative humidity was maintained at 70%. Approximately 2 μg ml−1 of the IL60 and PpEBB1-IL60 plasmids was introduced into peach buds as the control and treatment condition, respectively.
Identification and phylogenetic analysis of PpEBB1
The most current nectarine protein and CDSs were downloaded from the JGI database (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Pprsica). To study the phylogenetic relationship between PpEBB1 and homologous genes from other plant species, a phylogenetic tree was created via MEGA 5.05 software, and the full-length protein sequences were subjected to multiple alignment by the neighbor-joining method of ClustalW; the bootstrap test was replicated 1000 times.
Total RNA extraction and quantitative real-time PCR-based experiments
Total RNA was isolated via an RNAprep Pure Plant Kit (polysaccharide- and polyphenolic-rich) (Tiangen, Beijing, China). The quality and quantity of the RNA were determined via 1.0% agarose gel electrophoresis and a NanoPhotometer P360 (Implen, Munich, Germany). Reverse transcription was performed with a PrimeScript RT Reagent Kit and gDNA Eraser (Perfect Real Time) (Takara Biotechnology, Dalian, China). The primers used for qRT-PCR are listed in Supplementary Table S1 at JXB online. SYBR Premix Ex Taq (Takara Biotechnology) was used to perform quantitative real-time PCR (qRT-PCR). The thermal cycling conditions for qRT-PCR were as follows: predenaturation at 95 °C for 10 min followed by 40 cycles of denaturation at 95 °C for 5 s, annealing at 60 °C for 30 s, and extension. Each sample was replicated three times.
Vector construction and overexpression of PpEBB1 in poplar
The PpEBB1 gene was obtained via a pair of primers (forward, 5′-ATGGAAGAGGCATTTAGAAGG-3′ and reverse, 5′-GTTCTGGACCCTGGC-3′) with the additional restriction enzyme cutting sites of SalI and BamHI to allow for the direct cloning of the PpEBB1 gene into the pRI101 vector under the control of the CaMV35S promoter. The resulting 35S:PpEBB1 fusion construct was subsequently introduced into Agrobacterium tumefaciens strain GV3101.
Poplar (Populus trichocarpa) was used for heterologous transformation. Poplar seedlings were propagated by stem segments in aseptic glass bottles containing Murashige and Skoog (MS) media supplemented with 0.1 mg l−1 naphthylacetic acid (NAA). The poplar seedlings were subsequently grown at temperatures of 21–25 °C/15–18 °C (day/night) under a 12 h light (200 µmol m−2 s−1)/12 h dark photoperiod. Approximately 1 month later, approximately the fourth to sixth expanded leaves from the apex were used for injection. The leaves were cultivated on MS medium supplemented with 0.1 mg l−1 NAA, 0.2 mg l−1 6-benzylaminopurine, 0.01 mg l−1 thidiazuron, 800 mg l−1 timentin, and 50 mg l−1 kanamycin after injection. One or two months later, the small seedlings growing from the leaves were transferred to MS medium supplemented with 0.1 mg l−1 NAA and 50 mg l−1 kanamycin to grow roots. The screened transgenic lines were then transplanted into soil in a greenhouse.
RNA-sequencing data analysis
Populus trichocarpa genome annotations were downloaded from the JGI database (http://genome.jgi.doe.gov/Ptrichocarpa/download/_JAMO/587b0ae47ded5e4229d885bd/Ptrichocarpa_444_v3.1.gene_exons.gff3.gz?requestTime=1501142324). All reads were of high quality according to the distribution of the raw data. To identify differentially expressed genes (DEGs), DESeq2 v1.6.3 was used. Transcript abundance was normalized to fragments per kilobase of transcript per million mapped reads (FPKM) (Trapnell et al., 2010). The P-value and fold change were used to screen the DEGs, and genes with |log2 ratio|≥1 and q≤0.05 were identified as DEGs. DEGs were implemented by the hypergeometric test for Gene Ontology (GO, http://geneontology.org/) enrichment analysis. GO terms with q<0.05 were considered to be significantly enriched. The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of DEGs was implemented by a hypergeometric test, in which the P-value was adjusted by multiple comparisons, yielding a q-value. KEGG pathways with q<0.05 were considered significantly enriched.
Yeast two-hybrid assay
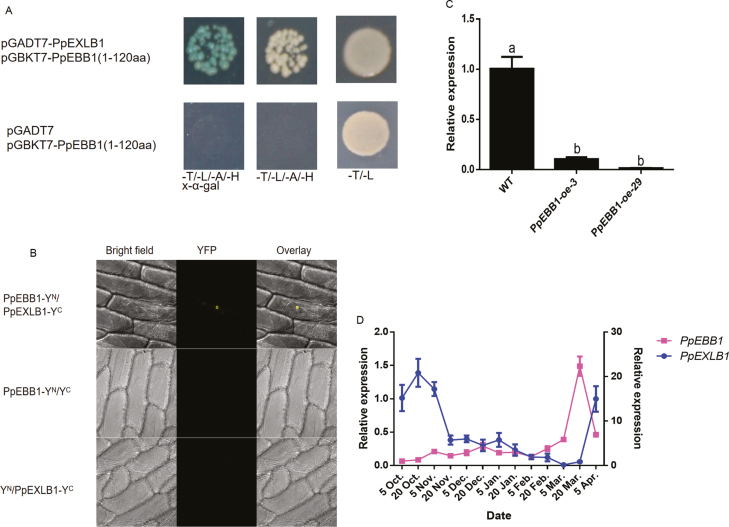
A yeast two-hybrid (Y2H) library was constructed with peach buds. PpEBB11−424 aa, PpEBB11−120 aa, and PpEBB1121−424 aa were cloned and inserted into a pGBKT7 vector as bait. The vectors were transformed into Y2H-competent cells and cultured at 30 °C on −Trp, −Trp/X-α-gal, and −Trp/X-α-gal/aureobasidin A (AbA) media. PpEXLB was cloned and inserted into a pGADT7 vector as prey. The bait and prey were subsequently transformed into Y2H-competent cells and cultured on −Trp/−Leu, −Leu/−Trp/−His/−Ade, and −Leu/−Trp/−His/−Ade media with X-α-gal.
Bimolecular fluorescence complementation assay
The CDSs of PpEBB1 and PpEXLB were transferred into pSPYNE and pSPYCE, respectively. The recombinant plasmids were then transformed into A. tumefaciens GV3101. The PpEBB1-pSPYNE and PpEXLB-pSPYCE plasmids were mixed together (1:1), after which onion epidermal cells were transfected with the mixture for 30 min; a mixture of PpEBB1-pSPYNE and pSPYCE was used as a control. The onion epidermal cells were then transferred to MS medium. Forty-eight hours later, the onion epidermal cells were observed at an excitation wavelength of 514 nm under a laser scanning confocal microscope (LSM880) (Carl Zeiss, Oberkochen, Germany).
Hormone determination
Samples (approximately 0.5 g of fresh weight per sample) were taken from PpEBB1-oe plants and wild-type (WT) and immersed immediately in liquid nitrogen. The contents of endogenous plant hormones were determined according to the method of Cai et al. (2015). The experiment was repeated at least three times for each sample.
Statistical analysis
Charts were constructed with GraphPad Prism software. Statistical analysis was performed with IBM SPSS Statistics 19 software, and Duncan’s test was used to analyse the data. A significance level of P<0.05 was applied.
Results
PpEBB1 may participate in bud break
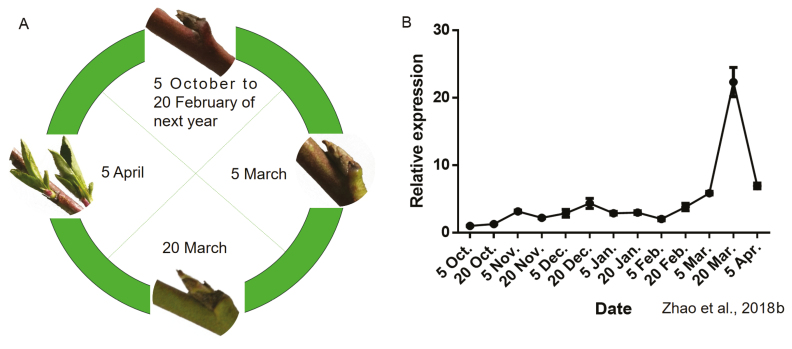
Peach buds were wrapped in thick bud scales and displayed no visible growth from 5 October 2016 to 20 February 2017. On 5 March, bud growth was obvious. The buds began to break away from the bud scales on 20 March, and new leaves grew outward on 5 April (Fig. 1A). In our previous study, we identified a gene named PpEBB1 that might function in the bud break of peach. The relative expression of PpEBB1 was very low and nearly constant during dormancy, whereas it began to be up-regulated on 20 February, peaked on 20 March, then decreased rapidly on 5 April, consistent with the date of bud break (Fig. 1B) (Zhao et al., 2018b).
Fig. 1.
Morphology of peach buds and the relative expression of PpEBB1. (A) Morphology of peach buds during the various stages of dormancy and bud break from 5 October 2016 to 5 April 2017. (B) Expression level of PpEBB1 during the various stages of dormancy and bud break from 5 October 2016 to 5 April 2017. The values represent the means ±SD of three replicates.
PpEBB1 can promote bud break
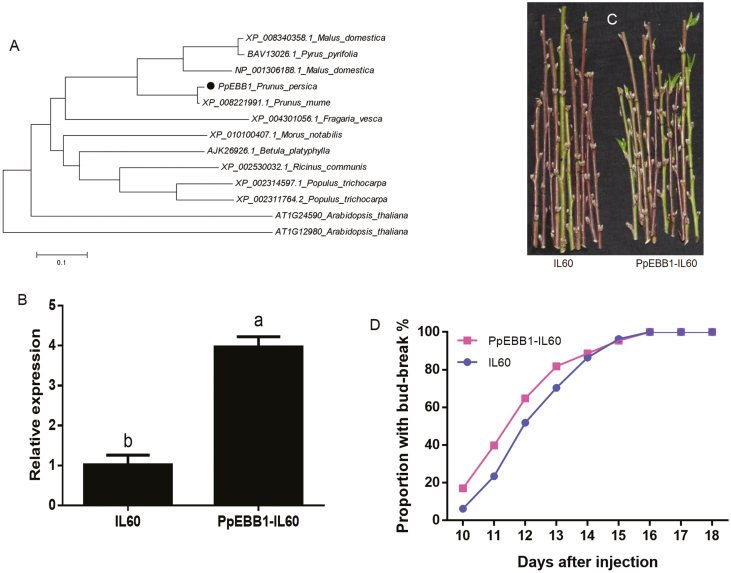
Phylogenetic analysis revealed that PpEBB1 was highly homologous to EBB1 in apple and pear (Fig. 2A), which may function in bud break because of its up-regulated expression when buds began to regrow. To elucidate the function of PpEBB1 in bud break, the PpEBB1 gene was cloned, and a PpEBB1-IL60 construct was generated; the construct was transformed into dormant peach buds, and an empty IL60 vector was used as a control. The transcript level of PpEBB1 significantly increased in PpEBB1-IL60-transfected buds compared with control buds (Fig. 2B), and the date of bud break was earlier for the buds transfected with PpEBB1 than for the buds transfected with the control (Fig. 2C, D).
Fig. 2.
Transient expression of PpEBB1 in peach buds. (A) Unrooted neighbor-joining tree of orthologous genes of PpEBB1 from different species. (B) Transient conversion of PpEBB1 in peach buds with an EBB1 overexpression level three times greater than that in the control. The values represent the means ±SD of three replicates, and the different letters above the bars represent significant differences; P<0.05. (C, D) Comparison of peach buds injected with a PpEBB1-IL60 vector and an empty IL60 vector (as a control). Ninety buds were divided into three groups as three replicates for transformation in each treatment. The proportion of bud break was calculated according to the sum of the buds in the three groups, and the buds that ultimately burst were included in the total buds.
Moreover, transgenic poplar plants overexpressing PpEBB1 (PpEBB1-oe) (Supplementary Fig. S1) were screened and planted in the greenhouse. The PpEBB1-oe plants (Fig. 3D, E) reactivated growth in early spring on 8 March when the WT plants remained dormant (Fig. 3A–C). Thus, regrowth occurred earlier in the transgenic PpEBB1-oe plants than in the WT plants. These results indicated that PpEBB1 plays important roles in promoting bud break.
Fig. 3.
Heterologous transformation of PpEBB1 induces early bud burst in poplar. Transgenic plants were planted in a greenhouse. Images were taken on 8 March 2019. (A–C) are WT plants, (D, E) are transgenic plants.
GO and KEGG analyses
To explore how PpEBB1 acts on bud break, RNA-seq was used to study the transcriptomic changes in the EBB1-oe poplar plants. A total of 2534 DEGs were identified (Supplementary Dataset S1), including 1525 genes whose expression was up-regulated and 1009 genes whose expression was down-regulated in the PpEBB1-oe plants compared with the WT plants. The transcriptomic data were successfully validated by qRT-PCR for a subset of 15 DEGs (Supplementary Fig. S2). DEGs were subjected to KEGG analysis, which highlighted several pathways, including the plant hormone signal transduction, zeatin biosynthesis, and tryptophan metabolism pathways (Supplementary Dataset S2). DEGs were also subjected to GO analysis and were classified into three categories: biological processes, cellular components, and molecular functions (data not shown). We mainly focused on GO terms associated with biological processes (Supplementary Dataset S3). From these GO terms, genes involved in hormone metabolism, especially that of auxin and cytokinin, the cell cycle process, and cell wall modification, which are processes that also function in dormancy, were significantly enriched; therefore, these genes were selected for further analysis.
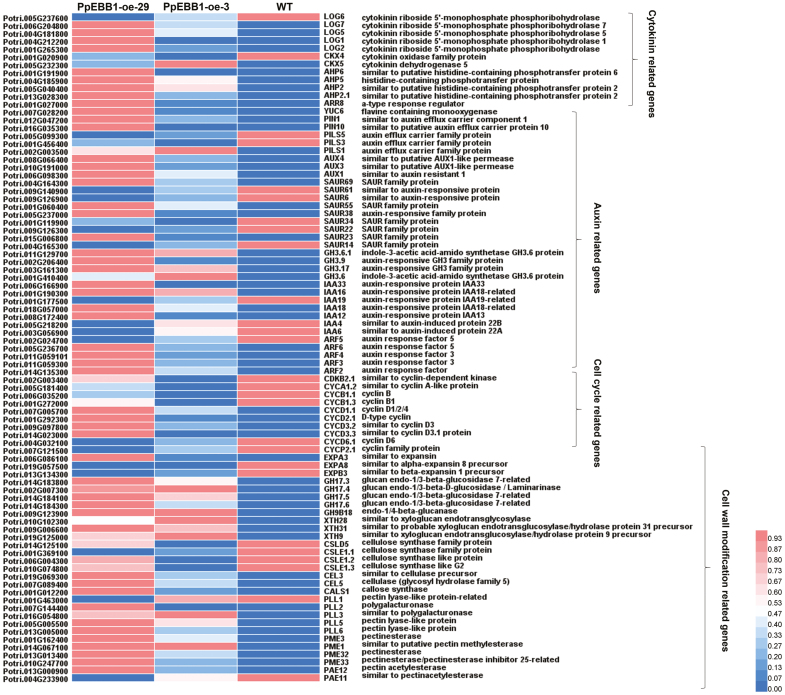
DEGs related to cytokinin and auxin metabolism with the overexpression of PpEBB1
Many DEGs associated with auxin and cytokinin metabolism, the cell cycle process and cell wall modification were selected from the transcriptomic data, and the relative expression of these genes in EBB1-oe plants and WT plants was assessed by qRT-PCR (Supplementary Dataset S4). A heat map (Fig. 4) was formed from the qRT-PCR data. Many genes associated with cytokinin and auxin metabolism showed marked changes in expression in the comparison of EBB1-oe and WT plants (Fig. 4; Supplementary Dataset S4). Among the DEGs associated with cytokinin biosynthesis, which were mainly lonely guy (LOG) and dehydrogenase-like cytokinin oxidase/dehydrogenase (CKX) genes, the expression levels of most were up-regulated in the EBB1-oe plants, except for LOG6 and CKX4. Regarding the cytokinin signaling component, there were four Arabidopsis histidine phosphotransfer proteins (AHPs) among the DEGs, and the expression level of all of these genes was higher in the transgenic plants than in the WT plants. The expression of ARR8, a type-A Arabidopsis response regulator, was up-regulated in the transgenic plants in our study. These results indicated that PpEBB1 could regulate cytokinins through the cytokinin biosynthesis pathway and signaling response.
Fig. 4.
Heat map of the DEGs between WT and transgenic (PpEBB1-oe-3 and PpEBB1-oe-29) poplar plants mainly included cytokinin and auxin metabolism, cell cycle, and cell wall modification-related genes. The annotation of each gene was retrieved from https://phytozome.jgi.doe.gov/pz/portal.html#, and the abbreviated name is provided for reference. The qRT-PCR data for the heat map are the means of three replicates.
For the auxin metabolism pathway, an auxin biosynthesis gene, YUCCA 6 (YUC6), was up-regulated in the EBB1-oe plants. The expression levels of PIN-formed genes (PINs) and auxin-resistant mutation genes (AUXs), which encode auxin efflux and influx carrier proteins, also changed. In addition, genes involved in auxin signaling, including members of the small auxin-up RNA (SAUR), GH3 and auxin/indole-3-acetic acid (Aux/IAA) family, were identified, and nine, four, and seven genes in the SAUR, GH3, and Aux/IAA families, respectively, were differentially expressed. The expression levels of four and one auxin response factor genes (ARFs) were up-regulated and down-regulated, respectively. These results indicated that PpEBB1 can regulate auxin through auxin biosynthesis, transport, and signaling responses.
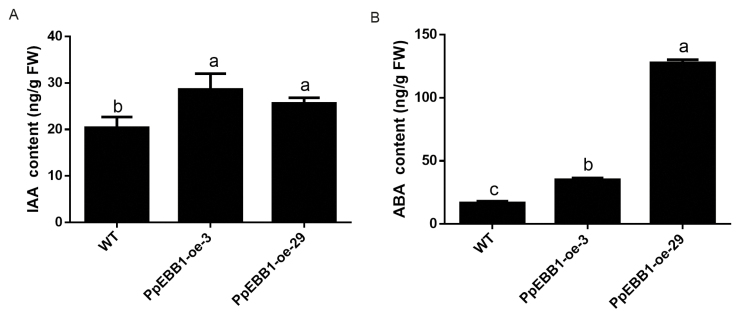
Owing to the important effect of PpEBB1 on cytokinin- and auxin-related genes, the levels of indoleacetic acid (IAA) and cytokinins were measured. IAA significantly increased in the transgenic plants compared with the WT plants (Fig. 5A). Six types of cytokinins were detected, and only the trans-zeatin (tZ) content was higher in the PpEBB1-oe plants than in the WT plants. The total tZ-type cytokinin was higher in the PpEBB1-oe plants than in the WT plants, while the total N6-(Δ 2-isopentenyl)-adenine (iP)-type cytokinin was lower (Table 1). In addition to cytokinin and auxin, some genes related to GA and ABA were also differentially expressed according to the transcriptomic data. Owing to the important role of these hormones during dormancy and bud break, the content of these hormones was also determined. The results showed that the ABA content increased in the PpEBB1-oe plants (Fig. 5B), which was unexpected. Three types of GA were detected: GA1 and GA4 were detected only in the WT plants, and GA3 was detected only in the PpEBB1-oe plants (Table 2).
Fig. 5.
Hormone content in the WT and transgenic plants. (A) Content of IAA; (B) content of ABA. The values represent the means ±SD of three replicates, and the different letters above the bars represent significant differences; P<0.05.
Table 1.
Levels of different types of cytokinin in PpEBB1-oe plants
| Sample | iP | iPR | tZ | tZR | cZ | cZR | Total tZ-types | Total iP-types |
|---|---|---|---|---|---|---|---|---|
| WT | 0.09±0.01 | 21.34±0.25 | 0.25±0.02 | 0.22±0.01 | 0.27±0.02 | 17.33±0.73 | 0.47±0.03 | 21.43±0.25 |
| PpEBB1-oe-3 | 0.05±0.00 | 5.26±0.04 | 0.36±0.04 | 0.43±0.02 | 0.30±0.03 | 4.77±0.24 | 0.79±0.06 | 5.31±0.04 |
| PpEBB1-oe-29 | 0.04±0.00 | 12.00±0.42 | 0.13±0.01 | 1.32±0.11 | 0.05±0.01 | 6.48±0.09 | 1.44±0.12 | 12.04±0.42 |
Values shown are means ±SD (ng g−1). cZ, cis-zeatin; cZR, cZ riboside; iP, N6-(Δ 2-isopentenyl)-adenine; iPR, iP riboside; tZ, trans-zeatin; tZR, tZ riboside.
Table 2.
Levels of different types of GA in PpEBB1-oe plants
| Sample | GA1 | GA3 | GA4 |
|---|---|---|---|
| WT | 0.67±0.01 | n.d. | 0.08±0.00 |
| PpEBB1-oe-3 | n.d. | 0.11±0.01 | n.d. |
| PpEBB1-oe-29 | n.d. | 0.23±0.01 | n.d. |
Values shown are means ±SD (ng g−1). n.d., not detected.
DEGs related to the cell cycle and cell wall modification with the overexpression of PpEBB1
DEGs related to the cell cycle and cell wall modification were detected (Fig. 4; Supplementary Dataset S4). Many CYC genes were differentially expressed, such as CYCD1.1, CYCD2.1, and CYCD3.2/3.3, whose expression was up-regulated, while the expression levels of CYCA1.2, CYCB1.1/1.3, CYCD6.1, and CYCP2.1 were down-regulated. The expression of a CDK-encoding gene, CDKB2, was down-regulated in the transgenic plants compared with the WT plants. A large number of the DEGs associated with expansins and the metabolism of cell wall components were identified on the basis of the GO annotations. The heat map showed that the expression levels of three expansin, five glycosyl hydrolase, three xyloglucan endotransglucosylase/hydrolase, six cellulose, one callose, and 11 pectin metabolism-related genes were altered (Fig. 4; Supplementary Dataset S4). These results indicated that PpEBB1 plays a crucial role in the cell cycle and cell wall modification.
PpEBB1 interacts with PpEXLB1
To examine how PpEBB1 is involved in bud break and to identify the genes that may interact with PpEBB1, a Y2H library was constructed from buds collected from dormancy to bud break. The N-terminus (1–120 amino acids (aa)) of PpEBB1 was used for screening interacting genes whose products contain an AP2/ERF domain, as the full-length protein is autoactivated (Supplementary Fig. S3). Ten genes (Supplementary Table S2) were screened and subsequently tested for their ability to interact with PpEBB1. The full-length cDNA of these genes was inserted into a pGADT7 vector as prey and together with BD-EBB11−120aa was used for Y2H assays. The expansin-like B1 (EXLB1; Prupe.5G057900) gene was found to interact with the N-terminal AP2/ERF domain (1–120 aa) of EBB1 (Fig. 6A), which might function in loosening the cell wall to regulate cell expansion. The interaction between PpEBB1 and PpEXLB was also verified by bimolecular fluorescence complementation (BiFC) assays (Fig. 6B).
Fig. 6.
PpEBB1 interacts with PpEXLB, and the expression pattern of PpEXLB during the dormancy stage. (A) Interaction between PpEBB1 and PpEXLB in a Y2H assay. Because of the autoactivation of the full-length PpEBB1 protein, we used aa 1–120 to perform the experiment. (B) Interaction between PpEBB1 and PpEXLB in a BiFC assay. (C) Compared with those in the WT plants, the expression levels of PpEXLB1 in two lines of PpEBB1-oe poplar decreased. (D) Expression level of PpEXLB1 during different dormancy stages from 5 October 2016 to 5 April 2017; blue represents PpEXLB1 (left axis), and purple represents PpEBB1 (right axis). The values represent the means ±SD of three replicates, and the different letters above the bars represent significant differences; P<0.05.
In PpEBB1 transgenic plants, the relative expression of PpEXLB1 was down-regulated (Fig. 6C). The expression levels of PpEXLB1 in peach buds were also investigated in parallel with PpEBB1 from late autumn to early spring. In the growth cessation stage, the expression level of PpEXLB1 was high, while the level was decreased in the dormancy stage and was lowest on 5 March, the time at which the expression of PpEBB1 was increasing. The expression of PpEXLB was then rapidly up-regulated on 5 April, which was the opposite of the expression pattern of PpEBB1 (Fig. 6D).
Discussion
The EBB1 gene in peach was identified; it has high sequence homology with those of apple, pear, and poplar (Fig. 2A). The relative expression of EBB1 during the dormancy period in apple (Wisniewski et al. 2015) and pear (Anh Tuan et al., 2016) was similar to that in peach in our study (Fig. 1), indicating that EBB1 might be associated with bud break. The results for both transformed peach buds and transgenic poplar (Figs 2C, D, 3) strongly confirmed that PpEBB1 plays an important role in the resumption of growth after winter dormancy. First, the expression level of EBB1 in buds was very low during the majority of the dormancy period but sharply increased at bud break, after which it rapidly decreased. Second, the overexpression of PpEBB1 was sufficient to accelerate bud burst. Third, these results were consistent with those of studies on apple, pear, and poplar (Yordanov et al., 2014; Wisniewski et al. 2015; Anh Tuan et al., 2016). Therefore, the role of EBB1 in promoting bud break is important and relatively conserved across perennial woody plant species.
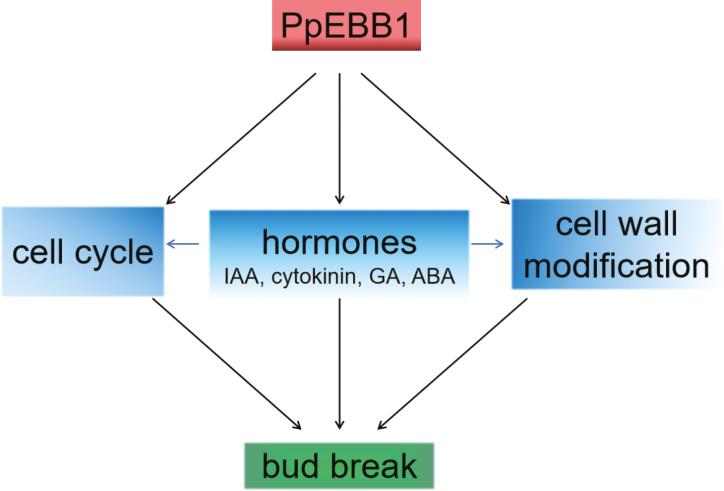
The molecular mechanism by which EBB1 regulates bud break remains unclear; thus, we used RNA-seq to study the transcriptomic changes in PpEBB1-oe poplar plants. GO analysis of the DEGs revealed terms associated with many processes critical for bud break, including hormone metabolism, the cell cycle, and cell wall modification (Supplementary Dataset S3), suggesting that PpEBB1 might promote bud break by regulating the metabolism of hormones, the cell cycle, and cell wall modification.
Among the DEGs associated with hormones, we focused mainly on genes related to cytokinin and auxin because genes involved in the metabolism of these two hormones were more enriched according to the GO analysis (Supplementary Dataset S3). Cytokinin is known to promote bud break. A previous study (Roman et al., 2016) in rose showed that cytokinin was the key factor that controlled bud outgrowth due to the exogenous supply of synthetic cytokinin 6-benzylaminopurine, which led to bud outgrowth in the dark, whereas two inhibitors of cytokinin perception repressed bud outgrowth, demonstrating the significant role of cytokinin in bud outgrowth. In Arabidopsis, cytokinin plays an important role in the formation of the shoot apical meristem (SAM), as the content of cytokinin causes changes in the SAM (Tokunaga et al., 2012). Moreover, in the SAM, the expression of WUSCHEL (WUS) is positively regulated by cytokinin (Gordon et al., 2007). Recent studies have shown that type-B ARRs can directly bind to the promoter region of WUS to promote its expression (Wybouw and De Rybel, 2019). Among the different types of cytokinin, tZ and iP generally presented relatively high activity, while cis-zeatin (cZ) presented relatively low or no activity (Sakakibara, 2006). In our study, overexpression of PpEBB1 improved the total content of tZ-type cytokinin and reduced the total iP-type cytokinin (Table 1). A study in rose showed that light-induced cytokinin accumulation in buds preferentially induced the tZ-type compared with the iP-type cytokinin (Roman et al., 2016). The levels of tZ and iP are balanced in plants by the antagonistic regulation of cytokinin-related enzymes, which interact with other factors, such as auxin and ABA, to regulate plant responses (Sakakibara, 2006). In addition, exogenous application of cytokinin can accelerate bud burst in rose, perhaps by inducing mechanisms closely related to germination, such as the expression of sugar-, IAA- and cell cycle-related genes (Roman et al., 2016). Cytokinin was shown to be capable of regulating cell division by interacting with CYCD3 and controlling the G2–M and G1–S transitions in the cell cycle (Zhang et al., 2005; Dewitte et al., 2007; Schaller et al., 2014). These results confirm that the changes in cytokinin levels influenced by PpEBB1 may act as a factor in promoting bud break or by regulating other bud break-related genes and metabolic pathways indirectly (Fig. 7).
Fig. 7.
Hypothetical model of PpEBB1 function in bud break. PpEBB1 can regulate genes associated with hormones, the cell cycle and cell wall modifications to promote bud break. In addition to accelerating bud break directly, hormones may act by regulating the cell cycle and cell wall modifications.
A study (Yordanov et al., 2014) of EBB1 in poplar suggested a possible connection between EBB1 and auxin signaling because genes involved in auxin signaling were enriched in EBB1-modified transgenics, and the orthologous gene DRN/DRNL in Arabidopsis can act via the auxin signal transduction pathway (Cole et al., 2009). DEGs related to auxin synthesis and transport and auxin signaling pathways were significantly enriched in PpEBB1-oe poplar plants in our study. The up-regulated expression of the auxin synthesis-related gene YUC6 was strongly correlated with an increased IAA content in the transgenic plants in this study (Figs 4, 5A; Supplementary Dataset S4). Although it is well known that auxin primarily plays a significant role in paradormancy (Balla et al., 2011), the function of auxin in bud break is not clear. By using qRT-PCR, we found that the expression levels of a large number of genes associated with auxin signaling pathways were altered (Fig. 4; Supplementary Dataset S4), indicating that EBB1 plays a very important role in auxin signaling. The auxin efflux carrier PIN3 was important for axillary bud outgrowth and could be directly repressed by BRC1 in cucumber (Cucumis sativus L.) (Shen et al., 2019). In our study, the expression of two PIN genes was up-regulated in the EBB1-oe plants, suggesting that EBB1 may regulate the transport of auxin. PIN protein polarization at the plasma membrane could also be promoted by cytokinin in Arabidopsis, resulting in bud outgrowth due to auxin export from buds (Waldie and Leyser, 2018). ABA is widely considered to be involved in dormancy induction and bud outgrowth inhibition, while GA breaks dormancy and promotes bud break. However, in our experiment, the EBB1-oe poplar plants, which displayed early bud break, presented increased ABA levels, and the content of GA1, GA3, and GA4 was completely different between the PpEBB1-oe plants and the WT plants (Table 2), indicating that the levels of a particular hormone, such as high levels of ABA, have a limited effect on dormancy and bud break. Sechet et al. (2016) also reported that early germination is not always accompanied by low ABA and high GA contents in Arabidopsis seeds. These results indicate that bud break may require a balance of various hormones rather than depending on one hormone; this is currently unclear.
Vegetative bud growth following dormancy release is often accompanied by increased cell division. DRN/DRNL affects cell cycle progression and provides local competence for the G1–S transition in the SAM of Arabidopsis (Seeliger et al., 2016). DEGs associated with the cell cycle, mainly CYCs and CDKs, were identified in our study (Fig. 4; Supplementary Dataset S4). These results confirmed that EBB1 has a significant influence on cell division by regulating the expression of cell cycle-related genes, which may be one of the reasons EBB1 promotes bud break. A study (Anh Tuan et al., 2016) of EBB in pear reported that EBB could activate the expression of CYCD3 genes by directly binding to their promoter or by interacting with other transcription factors first, after which the resulting complex increased the activities of the CYCD3 promoters, or the activity of the CYCD3 promoter could be enhanced by an intermediate transcription factor, which could be up-regulated by EBB. In addition, the cell cycle could be regulated directly by plant hormones. Auxin stimulates the G1–S transition of the cell cycle to promote lateral root initiation, and cytokinin interacts with CYCD3 in shoot tissue (Zhang et al., 2005; Dewitte et al., 2007); CYCB and CDKB could be regulated by hormones such as auxin, cytokinin and GA (Francis and Sorrell, 2001). Hormones including ABA, GA, auxin, and cytokinin have distinct functions in the regulation of the cell cycle (Francis and Sorrell, 2001). Therefore, EBB1 could regulate bud break by directly activating cell cycle regulatory genes or by indirectly mediating cell cycle-related genes via hormones (Fig. 7), facilitating the reinitiation of cell division and inducing bud break.
Research on the role of the cell wall in the bud break of perennial woody plants is rare. Microarray analyses showed that cell wall remodeling processes play important roles in germination in Arabidopsis (Penfield et al., 2006; Morris et al., 2011; Endo et al., 2012; Dekkers et al., 2013). The expression levels of genes associated with cell wall modification, including those involved in the metabolism of pectin, cellulose, hemicellulose, glycosyl, callose, and expansins, changed significantly when EBB1 was overexpressed in our study (Fig. 4; Supplementary Dataset S4). Cell wall modifications, such as pectin modification and activity of expansins and XTH, have been proposed to regulate the seed-to-seedling transition (Bassel, 2016). Loss of function of AtXTH31/XTR8 promotes germination in Arabidopsis (Endo et al., 2012). Sechet et al. (2016) also reported that xyl1 mutants showed reduced dormancy and could germinate on media lacking gibberellin. Pectin demethyl-esterification regulated by PME is thought to regulate cell expansion (Bordenave and Goldberg, 1994). Pectin demethyl-esterification and callose were found to be associated with bud development and played a role in growth–dormancy cycling by influencing cell wall permeability in Norway spruce (Lee et al., 2017). These results demonstrated that the cell wall might also be important for the bud break of perennial woody plants, confirming that cell wall modification might be one of the ways EBB1 influences bud break. Our Y2H and BiFC assay investigating the interaction between PpEBB1 and PpEXLB (Fig. 6A, B) further clarified that PpEBB1 could regulate cell wall remodeling by directly interacting with cell wall modification-related genes. A study on rose (Roman et al., 2016) showed that the expression of RhEXP increased during bud outgrowth and that the activation of the organogenic activity of SAM in response to cytokinin appeared to be mediated by RhEXP. In addition, cell wall modification could also be regulated by hormones. Previous research showed that activity of a pectate lyase could be induced by auxin in Zinnia elegans (Domingo et al., 1998). GO enrichment analysis of excess and deficient levels of cytokinin revealed several cytokinin-related genes involved in cell wall modification (Maruyama-Nakashita et al., 2004; Hawkesford and Kok, 2006; Takahashi et al., 2011; Skalák et al., 2019). Recently, a study investigating the interplay between cytokinin and expansins in Arabidopsis roots revealed that EXPA1 could be activated by ARR1 (Pacifici et al., 2018). On the basis of our results, EBB1 directly regulated cell wall modification by interacting with related proteins, such as PpEXLB, or indirectly regulated this process by first mediating hormone levels (Fig. 7).
Conclusion
The EBB1 gene is involved in the acceleration of bud break, and this function is conserved in perennial woody plants. Understanding the mechanism by which EBB1 functions in bud break can provide a new way for dormancy-related molecular breeding and genetic engineering to be used to develop plants that can avoid bud damage. The results of our study suggest that EBB1 promotes bud break by regulating various mechanisms, such as hormones, cell division, and cell wall modification, and these factors operate synergistically to promote bud break (Fig. 7).
Supplementary data
Supplementary data are available at JXB online.
Dateset S1. DEGs in EBB1-oe lines compared with WT.
Dateset S2. KEGG pathways identified by differentially expressed genes in EBB1-oe poplar.
Dateset S3. Ontology classification associated with BP of differentially expressed genes in EBB1-oe poplar.
Dateset S4. Relative expression of genes are associated with auxin and cytokinin metabolism, cell cycle, and cell wall modification (data presented are the means of three replicates).
Fig. S1. Identification of transgenic poplar lines.
Fig. S2. Verification of RNA-seq results via qRT-PCR.
Fig. S3. Autoactivation of the full-length PpEBB1, PpEBB11−120aa, and PpEBB1120−424aa sequences by a Y2H assay.
Table S1. Primers used for qRT-PCR in this study.
Table S2. Genes that may interact with PpEBB1, as screened by Y2H.
Acknowledgements
This study was funded by the National Key Research and Developmental Program of China (2018YFD1000104) and the Shandong Province Modern Agricultural Technology System Fruit Innovation Team. We thank Dr Yujin Hao (Shandong Agricultural University, China) for providing the IL60 vector.
Author contributions
DG, XF and XZ designed the study; XZ, XH, QW, XW, XC and LL performed the experiments and analysed the data; XZ wrote the paper.
References
- Aksenova NP, Sergeeva LI, Konstantinova TN, Golyanovskaya SA, Kolachevskaya OO, Romanov GA. 2013. Regulation of potato tuber dormancy and sprouting. Russian Journal of Plant Physiology 60, 301–312. [Google Scholar]
- Anh Tuan P, Bai S, Saito T, Imai T, Ito A, Moriguchi T. 2016. Involvement of EARLY BUD-BREAK, an AP2/ERF transcription factor gene, in bud break in Japanese pear (Pyrus pyrifolia Nakai) lateral flower buds: expression, histone modifications and possible target genes. Plant & Cell Physiology 57, 1038–1047. [DOI] [PubMed] [Google Scholar]
- Balla J, Kalousek P, Reinöhl V, Friml J, Procházka S. 2011. Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. The Plant Journal 65, 571–577. [DOI] [PubMed] [Google Scholar]
- Barbier FF, Dun EA, Kerr SC, Chabikwa TG, Beveridge CA. 2019. An update on the signals controlling shoot branching. Trends in Plant Science 24, 220–236. [DOI] [PubMed] [Google Scholar]
- Bassel GW. 2016. To grow or not to grow? Trends in Plant Science 21, 498–505. [DOI] [PubMed] [Google Scholar]
- Bielenberg DG, Wang Y, Fan S, Reighard GL, Scorza R, Abbott AG. 2004. A deletion affecting several gene candidates is present in the Evergrowing peach mutant. The Journal of heredity 95, 436–444. [DOI] [PubMed] [Google Scholar]
- Bielenberg DG, Wang Y, Li Z, Zhebentyayeva T, Fan S, Reighard GL, Scorza R, Abbott AG. 2008. Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genetics & Genomes 4, 495–507. [Google Scholar]
- Bordenave M, Goldberg R. 1994. Immobilized and free apoplastic pectinmethylesterases in mung bean hypocotyl. Plant Physiology 106, 1151–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busov V, Carneros E, Yakovlev I. 2016. EARLY BUD-BREAK1 (EBB1) defines a conserved mechanism for control of bud-break in woody perennials. Plant Signaling and Behavior 11, e1073873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadman CS, Toorop PE, Hilhorst HW, Finch-Savage WE. 2006. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. The Plant Journal 46, 805–822. [DOI] [PubMed] [Google Scholar]
- Cai BD, Yin J, Hao YH, Li YN, Yuan BF, Feng YQ. 2015. Profiling of phytohormones in rice under elevated cadmium concentration levels by magnetic solid-phase extraction coupled with liquid chromatography tandem mass spectrometry. Journal of Chromatography A 1406, 78–86. [DOI] [PubMed] [Google Scholar]
- Castède S, Campoy JA, Le Dantec L, Quero-García J, Barreneche T, Wenden B, Dirlewanger E. 2015. Mapping of candidate genes involved in bud dormancy and flowering time in sweet cherry (Prunus avium). PLoS ONE 10, e0143250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabikwa TG, Brewer PB, Beveridge CA. 2019. Initial bud outgrowth occurs independent of auxin flow from out of buds. Plant Physiology 179, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Tan Q, Sun M, Li D, Fu X, Chen X, Xiao W, Li L, Gao D. 2016. Genome-wide identification of WRKY family genes in peach and analysis of WRKY expression during bud dormancy. Molecular Genetics and Genomics 291, 1319–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M, Chandler J, Weijers D, Jacobs B, Comelli P, Werr W. 2009. DORNRÖSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136, 1643–1651. [DOI] [PubMed] [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Müller D, Domagalska MA, Leyser O. 2010. Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137, 2905–2913. [DOI] [PubMed] [Google Scholar]
- De Lorenzo G, Ferrari S, Giovannoni M, Mattei B, Cervone F. 2019. Cell wall traits that influence plant development, immunity, and bioconversion. The Plant Journal 97, 134–147. [DOI] [PubMed] [Google Scholar]
- Dekkers BJ, Pearce S, van Bolderen-Veldkamp RP, et al. . 2013. Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiology 163, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devitt ML, Stafstrom JP. 1995. Cell cycle regulation during growth-dormancy cycles in pea axillary buds. Plant Molecular Biology 29, 255–265. [DOI] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, et al. . 2007. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proceedings of the National Academy of Sciences, USA 104, 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierck R, De Keyser E, De Riek J, Dhooghe E, Van Huylenbroeck J, Prinsen E, Van Der Straeten D. 2016. Change in auxin and cytokinin levels coincides with altered expression of branching genes during axillary bud outgrowth in Chrysanthemum. PLoS ONE 11, e0161732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo C, Roberts K, Stacey NJ, Connerton I, Ruíz-Teran F, McCann MC. 1998. A pectate lyase from Zinnia elegans is auxin inducible. The Plant Journal 13, 17–28. [DOI] [PubMed] [Google Scholar]
- Endo A, Tatematsu K, Hanada K, et al. . 2012. Tissue-specific transcriptome analysis reveals cell wall metabolism, flavonol biosynthesis and defense responses are activated in the endosperm of germinating Arabidopsis thaliana seeds. Plant & Cell Physiology 53, 16–27. [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Beveridge CA. 2009. Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiology 149, 1929–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D, Sorrell DA. 2001. The interface between the cell cycle and plant growth regulators: a mini review. Plant Growth Regulation 33, 1–12. [Google Scholar]
- Gillespie LM, Volaire FA. 2017. Are winter and summer dormancy symmetrical seasonal adaptive strategies? The case of temperate herbaceous perennials. Annals of Botany 119, 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Heisler MG, Reddy GV, Ohno C, Das P, Meyerowitz EM. 2007. Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134, 3539–3548. [DOI] [PubMed] [Google Scholar]
- Hawkesford MJ, De Kok LJ. 2006. Managing sulphur metabolism in plants. Plant, Cell & Environment 29, 382–395. [DOI] [PubMed] [Google Scholar]
- Horvath DP, Anderson JV, Chao WS, Foley ME. 2003. Knowing when to grow: signals regulating bud dormancy. Trends in Plant Science 8, 534–540. [DOI] [PubMed] [Google Scholar]
- Horvath DP, Chao WS, Suttle JC, Thimmapuram J, Anderson JV. 2008. Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genomics 9, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GA. 1987. Dormancy: a new universal terminology. Hortscience 22, 817–820. [Google Scholar]
- Lee Y, Karunakaran C, Lahlali R, Liu X, Tanino KK, Olsen JE. 2017. Photoperiodic regulation of growth-dormancy cycling through induction of multiple bud-shoot barriers preventing water transport into the winter buds of Norway spruce. Frontiers in Plant Science 8, 2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H. 2004. A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. The Plant Journal 38, 779–789. [DOI] [PubMed] [Google Scholar]
- Maurya JP, Bhalerao RP. 2017. Photoperiod- and temperature-mediated control of growth cessation and dormancy in trees: a molecular perspective. Annals of Botany 120, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K, Linkies A, Müller K, Oracz K, Wang X, Lynn JR, Leubner-Metzger G, Finch-Savage WE. 2011. Regulation of seed germination in the close Arabidopsis relative Lepidium sativum: a global tissue-specific transcript analysis. Plant Physiology 155, 1851–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Zhao ML, Chen MS, Pan BZ, Tao YB, Xu ZF. 2017. Comparative transcriptome analysis of axillary buds in response to the shoot branching regulators gibberellin A3 and 6-benzyladenine in Jatropha curcas. Scientific Reports 7, 11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Q, Li J, Cai D, Qian M, Jia H, Bai S, Hussain S, Liu G, Teng Y, Zheng X. 2016. Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. Journal of Experimental Botany 67, 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifici E, Mambro RD, Ioio RD, Constantino P, Sabatini S. 2018. Acidic cell elongation drives cell differentiation in the Arabidopsis root. The EMBO Journal 37, e99134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. 2006. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. The Plant Cell 18, 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameau C, Bertheloot J, Leduc N, Andrieu B, Foucher F, Sakr S. 2014. Multiple pathways regulate shoot branching. Frontiers in Plant Science 5, 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman H, Girault T, Barbier F, et al. . 2016. Cytokinins are initial targets of light in the control of bud outgrowth. Plant Physiology 172, 489–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. 2006. Cytokinins: activity, biosynthesis, and translocation. Annual Review of Plant Biology 57, 431–449. [DOI] [PubMed] [Google Scholar]
- Sarkar P, Bosneaga E, Auer M. 2009. Plant cell walls throughout evolution: towards a molecular understanding of their design principles. Journal of Experimental Botany 60, 3615–3635. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Street IH, Kieber JJ. 2014. Cytokinin and the cell cycle. Current Opinion in Plant Biology 21, 7–15. [DOI] [PubMed] [Google Scholar]
- Sechet J, Frey A, Effroy-Cuzzi D, et al. . 2016. Xyloglucan metabolism differentially impacts the cell wall characteristics of the endosperm and embryo during Arabidopsis seed germination. Plant Physiology 170, 1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger I, Frerichs A, Glowa D, Velo L, Comelli P, Chandler JW, Werr W. 2016. The AP2-type transcription factors DORNRÖSCHEN and DORNRÖSCHEN‑LIKE promote G1/S transition. Molecular Genetics and Genomics 291, 1835–1849. [DOI] [PubMed] [Google Scholar]
- Shen J, Zhang Y, Ge D, Wang Z, Song W, Gu R, Che G, Cheng Z, Liu R, Zhang X. 2019. CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber. Proceedings of the National Academy of Sciences, USA 116, 17105–17114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim D, Ko JH, Kim WC, Wang Q, Keathley DE, Han KH. 2014. A molecular framework for seasonal growth-dormancy regulation in perennial plants. Horticulture Research 1, 14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S, Mori H. 2001. Control of outgrowth and dormancy in axillary buds. Plant Physiology 127, 1405–1413. [PMC free article] [PubMed] [Google Scholar]
- Shinohara N, Taylor C, Leyser O. 2013. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biology 11, e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalák J, Vercruyssen L, Claeys H, et al. . 2019. Multifaceted activity of cytokinin in leaf development shapes its size and structure in Arabidopsis. The Plant Journal 97, 805–824. [DOI] [PubMed] [Google Scholar]
- Sun MY, Fu XL, Tan QP, Liu L, Chen M, Zhu CY, Li L, Chen XD, Gao DS. 2016. Analysis of basic leucine zipper genes and their expression during bud dormancy in peach (Prunus persica). Plant Physiology and Biochemistry 104, 54–70. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. 2011. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annual Review of Plant Biology 62, 157–184. [DOI] [PubMed] [Google Scholar]
- Tokunaga H, Kojima M, Kuroha T, Ishida T, Sugimoto K, Kiba T, Sakakibara H. 2012. Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. The Plant Journal 69, 355–365. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velappan Y, Signorelli S, Considine MJ. 2017. Cell cycle arrest in plants: what distinguishes quiescence, dormancy and differentiated G1? Annals of Botany 120, 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara R, Noriega X, Aravena K, Prieto H, Pérez FJ. 2017. ABA represses the expression of cell cycle genes and may modulate the development of endodormancy in grapevine buds. Frontiers in Plant Science 8, 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldie T, Leyser O. 2018. Cytokinin targets auxin transport to promote shoot branching. Plant Physiology 177, 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski M, Norelli J, Artlip T. 2015. Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Frontiers in Plant Science 6, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wybouw B, De Rybel B. 2019. Cytokinin – a developing story. Trends in Plant Science 24, 177–185. [DOI] [PubMed] [Google Scholar]
- Yamane H, Ooka T, Jotatsu H, Hosaka Y, Sasaki R, Tao R. 2011. Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. Journal of Experimental Botany 62, 3481–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordanov YS, Ma C, Strauss SH, Busov VB. 2014. EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proceedings of the National Academy of Sciences, USA 111, 10001–10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Diederich L, John PC. 2005. The cytokinin requirement for cell division in cultured Nicotiana plumbaginifolia cells can be satisfied by yeast Cdc25 protein tyrosine phosphatase: implications for mechanisms of cytokinin response and plant development. Plant Physiology 137, 308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Zhou Y, Ahmad S, Xu Z, Li Y, Yang W, Cheng T, Wang J, Zhang Q. 2018a Comprehensive cloning of Prunus mume dormancy associated MADS-box genes and their response in flower bud development and dormancy. Frontiers in Plant Science 9, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wang Q, Li C, Chen X, Xiao W, Gao D, Fu X. 2018b Genome-wide identification of ethylene responsive factor (ERF) family genes in peach and screening of genes related to germination. Chinese Bulletin of Botany 53, 612–624. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.