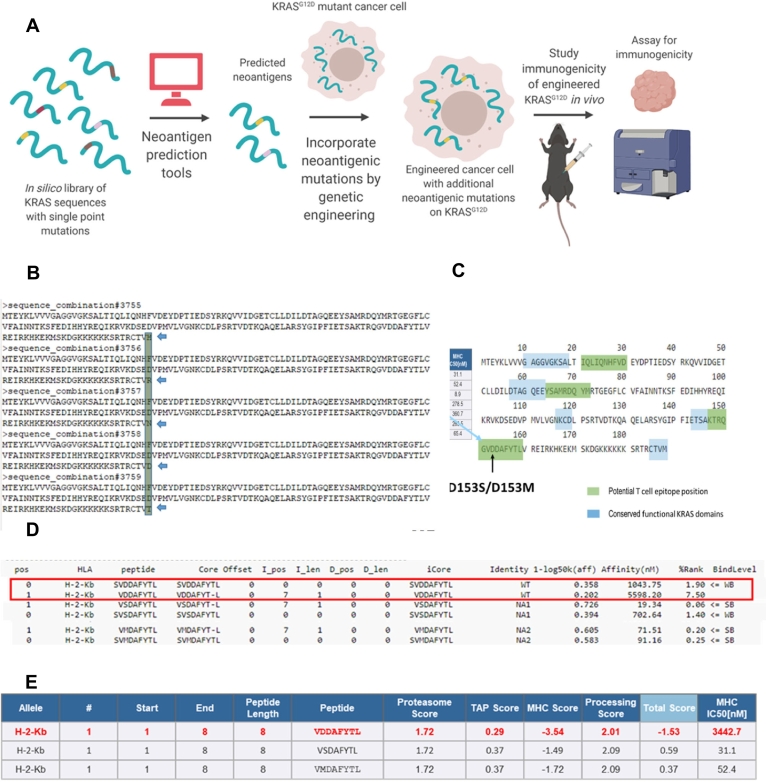
Fig. 1.
In silico prediction of neoantigens derivable from single point mutations on KRAS, a) Scheme of insertion of neoantigens via gene editing, b) Representative sequences from a library of mouse KRAS (UniProt P32883–2) sequences with single amino acid variations, c) Alignment of predicted mutation-derived neoantigens against conserved KRAS functional domains, d) Predicted changes in sequence scoring upon single point mutation, from weak binding level (WB) to strong binding level (SB), via NetMHC 4.0 Server, Technical University of Denmark, http://www.cbs.dtu.dk/services/NetMHC/, e) Predicted changes in sequence scoring upon single point mutation, via Immune Epitope Database and Analysis Resource, http://tools.iedb.org/main/tcell/