Abstract
Both autotaxin (ATX) and Forkhead Box M1 (FOXM1) have been commonly recognized as oncogenes in multiple types of human malignancies. However, the expression and biological functions of ATX in pancreatic ductal adenocarcinoma (PDAC), and its correlation with FOXM1 are poorly understood. The present study aimed to investigate their correlation and biological consequences in PDAC development. By dual luciferase reporter and chromatin immunoprecipitation assays, we found that ATX was a downstream transcriptional target gene of FOXM1. Further cellular functional experiments indicated that ATX was required for FOXM1-mediated PDAC cell proliferation and migration. Data from molecular biological experiments showed that ATX could enhance FOXM1 expression in turn by inhibiting the Hippo signaling pathway, suggesting that ATX and FOXM1 formed a positive feedback loop to facilitate PDAC progression. Using immunohistochemistry (IHC) method, both ATX and FOXM1 expression were found to be frequently up-regulated in PDAC tumor tissues when compared with adjacent normal tissues, and elevated ATX and FOXM1 expression were positively correlated with each other. In conclusion, the present work identified a positive feedback loop between ATX and FOXM1 which promotes PDAC cell proliferation and migration.
Keywords: ATX, FOXM1, pancreatic cancer, proliferation, migration
Introduction
As the most common type of pancreatic cancer, pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal human malignancies worldwide, with an extremely low 5-year survival rate [1,2]. Although big efforts have been made to improve the prognosis of patients with PDAC in the past decades, little improvement has been achieved due to the high tendency of PDAC to metastasize and resistance to chemoradiation therapy [3]. Therefore, a better understanding of the molecular mechanisms underlying the tumorigenesis and metastasis of PDAC is needed to identify novel potential therapeutic targets for the clinical management of PDAC.
Autotaxin (ATX), also known as nucleotide pyrophosphatase-phosphodiesterase 2 (ENPP2) [4], is a secreted glycoprotein that primarily catalyzes the hydrolysis of lysophosphatidylcholine to produce lysophosphatidic acid (LPA) [5], which is well-known to stimulate the proliferation, migration, and survival of many human malignancies [6,7]. High expression of ATX has been reported in different tumors and ATX overexpression contributes to tumor proliferation and invasion mainly through the induction of LPA [6,8,9]. In PDAC, increased serum ATX activity was found in patients with pancreatic cancer [10], and higher expression of ATX was observed in a highly invasive PANC-1 cell line [11]. Although these studies strongly suggest an aggressive role of ATX in PDAC development, its expression and biological functions in PDAC are largely unknown, and the molecular mechanisms that regulate ATX expression in PDAC as of yet remain unclear.
Forkhead box M1 (FOXM1) is a key member of the forkhead family of transcription factors [12,13]. Numerous studies have reported that FOXM1 plays a critical role in regulating cell cycle transition, cell proliferation and chromosome stability [13-16]. Like ATX, FOXM1 is also highly expressed in various types of human malignancies including PDAC [17,18], and overexpression of FOXM1 can stimulate tumor cell proliferation, invasion and angiogenesis in different tumors [17,19]. Although over-activation of both ATX and FOXM1 occur in many human cancers, it is unclear whether they are functionally correlated with each other. In the present study, we resorted to investigate their correlation and underlying molecular mechanisms in PDAC development.
Methods
Tissue microarray (TMA) and immunohistochemistry (IHC) analysis
For IHC analysis, a human PDAC TMA (#HLivHPanA150Su01) containing 60 tumor tissues and paired adjacent normal tissues as well as andditional 30 tumor tissues derived from 90 patients with PDAC was purchased from Shanghai Outdo Biotech CO., LTD., China. Standard IHC procedures were performed using specific antibodies against ATX (1:500, Abcam, #ab77104) and FOXM1 (1:200, Santa Cruz Biotechnology, #sc-502). The staining results were evaluated by two pathologists blinded to the clinical information independently according to the percentage of ATX/FOXM1-positive cells and staining intensity. Specifically, the percentage of ATX/FOXM1-positive cells was classified into 5 groups: < 10% (0), 10-25% (1), 25-50% (2), 50-75 (3), and > 75% (4). The staining intensity was divided into 4 groups: no staining (0), light brown (1), brown (2), and dark brown (3). Finally, the overall scores were calculated using the following formula: overall score = percentage score × intensity score. The samples with an overall score ≤ 6 were defined as weak, and > 6 were defined as strong.
Cell culture and reagents
Human PDAC cell lines PANC-1 and SW1990 were purchased from Shanghai Cell Bank of the Chinese Academy of Sciences Shanghai, China. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Corning, Inc., Corning, NY, USA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (M&C Gene Technology Ltd., Beijing, China). All of these cell lines were maintained in a humidified incubator at 37°C with 5% CO2. The Hippo signaling pathway (MST1/2) inhibitor XMU-MP-1 was purchased from Selleckchem, Houston, TX, USA.
Cell Transfection
The FOXM1-expressing plasmids pcDNA3.1-FOXM1 (isoform C) were gifts from Dr. Xie Keping (The University of Texas MD Anderson Cancer Center, Houston, TX, USA), and lentivirus carrying FOXM1-shRNA or FOXM1-ORF plasmids and the siRNAs against ATX (2’-O-methyl modified) were synthesized by GenePharma Co., Ltd., Shanghai, China. PDAC cells were transfected with plasmids or siRNAs using Lipofectamine 3000 (Invitrogen, USA) according to the protocol of the manufactures. To generate lentiviral stable cell lines, PDAC cells were infected with the lentiviruses described above with the assistance of polybrene (4 μg/ml), and puromycin (2 μg/ml, Sigma, MO, USA) was used to selected stably infected PDAC cells.
Western blots analysis
PDAC cells were lysed using a Radioimmunoprecipitation assay (RIPA) buffer supplemented with a protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) to extract total proteins. Standard western blot analysis was performed using primary antibodies anti-FOXM1 (1:200, Santa Cruz Biotechnology, #sc-502), anti-ATX (1:500, Abcam, #ab77104), and anti-β-actin (1:1000, Santa Cruz Biotechnology, #sc-81178), anti-mouse (Cell Signaling Technology) and anti-rabbit (Cell Signaling Technology) antibodies were used as secondary antibodies. The bands were visualized and quantified using a Li-Cor Odyssey protein imaging system (LI-COR Biosciences, Lincoln, NE, USA).
Quantitative real-time polymerase chain reaction (qPCR) analysis
Total RNAs were extracted from PDAC cells with Trizol reagent (Sigma-Aldrich, MO, USA) and quantified by a Fisher Scientific NanoDrop™ 2000 spectrophotometer (NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA) according to the manufacturer’s specifications, after which 1 μg RNA was reverse-transcribed into cDNA using the PrimeScript™ RT reagent kit (Takara Bio, Inc., Otsu, Japan). Standard qPCR analysis was performed with an ABI QuantStudioTM 6 Flex system (Thermo Fisher Scientific, Inc.) using SYBR green reagent (TaKaRa, Japan). Relative mRNA expression levels of FOXM1 and ATX were determined using the 2-ΔΔCt method and β-actin was used as an internal control. The following qPCR primers were used: For ATX, 5’-ACAACGAGGAGAGCTGCAAT-3’ (forward), 5’-AGAAGTCCAGGCTGGTGAGA-3’ (reverse); for FOXM1, 5’-GGGCGCACGGCGGAAGATGAA-3’ (Forward), 5’-CCACTCTTCCAAGGGAGGGCTC-3’ (Reverse); for β-actin: 5’-CCTGGCACCCAGCACAATG-3’ (forward), 5’-GGGCCGGACTCGTCATACT-3’ (reverse).
Colony formation assay
Briefly, PDAC cells were seeded into a 6-well plate at a density of 1 × 103 cells per well and then incubated for 2 weeks at 37°C with 5% CO2. 4% paraformaldehyde and crystal violet were used for the fixation and staining of the colonies, respectively. Next, the colonies were photographed, and the number of colonies was counted with image J software. Each experiment was performed in triplicate and repeated at least three times independently.
Cell proliferation assay
Cell viability of PDAC cells were evaluated using Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan) according to the manufacturer’s protocol. Briefly, the selected PDAC cells were seeded into a 96-well plate (3 × 103 cells/well) and incubated under normal conditions. At each scheduled time point (0, 24, 48, 72, 96 h), 10 μl of CCK-8 reagents were added into each well, and after incubation for 1 h at 37°C, the absorbance was measured using an automated microplate reader (SpectraMax M5, Molecular Devices LLC, CA, USA) at a wavelength of 450 nm.
Cell migration assay
Cell migration assay was conducted using a 24-well Transwell filter (pore size, 8 μm; Costar; Corning, Inc.). In brief, 800 μl DMEM containing 10% FBS was added to the bottom chamber, and 3 × 104 cells maintained in 400 μl DMEM without FBS were added into the upper chamber. After incubation under normal conditions for 24 h, the cells that stayed in the upper chamber were removed with a cotton swab, and the migrated cells were fixed in 4% paraformaldehyde for 15 min followed by staining with crystal violet for additional 15 min. Finally, five random fields were selected and the migrated cells were photographed and counted under an upright metallurgical microscope (Leica, Germany).
Dual luciferase reporter assay
To construct ATX promoter-activated luciferase reporter plasmid, wild or mutant ATX promoter was cloned into pGL3 basic luciferase reporter vector and the products were verified by DNA sequencing. A FOXM1 promoter-activated luciferase reporter plasmid containing 5’ FOXM1 sequences from -2430 to +66 bp relative to the transcription initiation site (designated as pFOXM1-2496) was constructed as described previously [20]. To conduct luciferase reporter assay, PDAC cells were seeded into a 24-well plate and transfected with the indicated promoter reporters. 24 h after transfection, the Dual-Glo® Luciferase Assay Kit (Promega, WI, USA) was used to evaluate the promoter activities of ATX or FOXM1 by quantifying both firefly and Renilla luciferase activity on a GloMax® 96 Microplate Luminometer (Promega, WI, USA).
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed on PANC-1 cells using a ChIP assay kit (Millipore, CA, USA) as per the manufacturer’s instructions. Briefly, an aliquot of the immunoprecipitated DNA samples was subjected to PCR and two primers were used to amplify the FOXM1-binding sites in the ATX promoter. Afterwards, the PCR products were resolved electrophoretically with a 2% agarose gel in 1 × TAE and the results were visualized using the ChemiDoc MP system (Bio-Rad Laboratories, Hercules, CA, USA). ChIP-qPCR analysis was performed using ChIP samples and SYBR green reagent (TaKaRa, Japan) according to the manufactures’ protocols. The primers used for ChIP assays were as follows: #1 site: 5’-AGCCATTGCACTCCAGCCT-3’ (forward) and 5’-CTGCTTGTTACCCAATATCC-3’ (reverse), CD81#2: 5’-GAATCTGTAATGAAACCCA-3’ (forward) and 5’-TGCGTAAACAAATACGAGAC-3’ (reverse).
Statistical analysis
Data was shown as mean ± standard error of the mean (SEM). Categorical data was analyzed using χ2 test, the significance of the data from independent experiments were determined using the two-tailed Student t-test or one-way analysis of variance (ANOVA), and the correlation between FOXM1 and ATX expression in PDAC tumor tissues was assessed using Pearson’s correlation. GraphPad Prism 7.0 software was used to perform statistical analysis. P values less than 0.05 were considered statistically significant.
Results
ATX expression is positively regulated by FOXM1 in PDAC cells
To evaluate the relationship between ATX and FOXM1 expression in PDAC, we generated stable PANC-1 and SW1990 cells with FOXM1 knockdown, and ATX expression was examined by western blots and qPCR analyses. As shown in Figure 1A and 1B, both protein and mRNA levels of ATX in PANC-1 and SW1990 cells were significantly decreased when FOXM1 was knocked down. On the contrary, ATX expression was enhanced in FOXM1 stably overexpressed PANC-1 and SW1990 cells (Figure 1C, 1D). These findings suggested that FOXM1 has a positive effect on ATX expression in PDAC cells.
Figure 1.
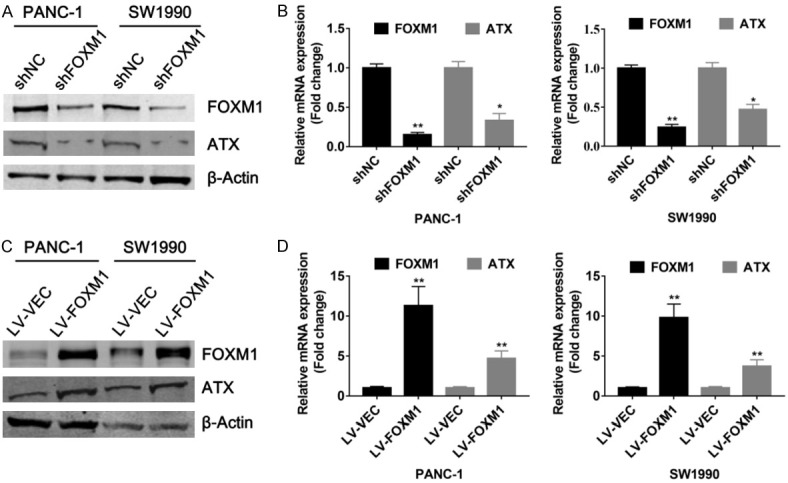
Regulation of ATX expression by FOXM1 in PDAC cells. (A and B) Proteins and RNAs were isolated from PANC-1 and SW1990 cells stably infected with LV-FOXM1 lentivirus or control lentivirus, and western blot (A) and qPCR (B) analyses were performed, respectively. (C and D) Proteins and RNAs were extracted from PANC-1 and SW1990 cells with stable expression of FOXM1, respectively. And western blot (C) and qPCR (D) analyses were carried out to determine the effects of FOXM1 on ATX expression. *P < 0.05, **P < 0.01.
FOXM1 is a transcriptional activator of ATX
Considering that FOXM1 functions as an important transcription factor, we next investigated whether FOXM1 regulates ATX expression at transcription level. -2 kb promoter region of human ATX gene was scanned to search potential FOXM1 DNA binding consensus sequence (5’-AT/CAAAT/CAA-3’), and two regions with putative FOXM1-binding sites were identified. Subsequently, we generated a series of luciferase reporter constructs containing wild-type (WT) and mutant (MUT) promoter region of ATX as presented in Figure 2A. The results of dual luciferase reporter assays indicated that FOXM1 knockdown decreased the promoter activity of WT-ATX, while no obvious effect was found on MUT-ATX promoter activity (Figure 2B). Concordantly, overexpression of FOXM1 significantly increased the WT-ATX but not MUT-ATX promoter activity in a dosage-dependent manner (Figure 2C), implying that FOXM1 regulates ATX expression via the putative FOXM1-binding sites. Furthermore, results of ChIP assay showed that the DNA fragments containing #1 site or #2 site sequence could be precipitated from PANC-1 cells with anti-FOXM1 antibody but not control IgG (Figure 2D, 2E), suggesting FOXM1 can directly bind to ATX promoter and activate ATX transcription in PDAC cells.
Figure 2.
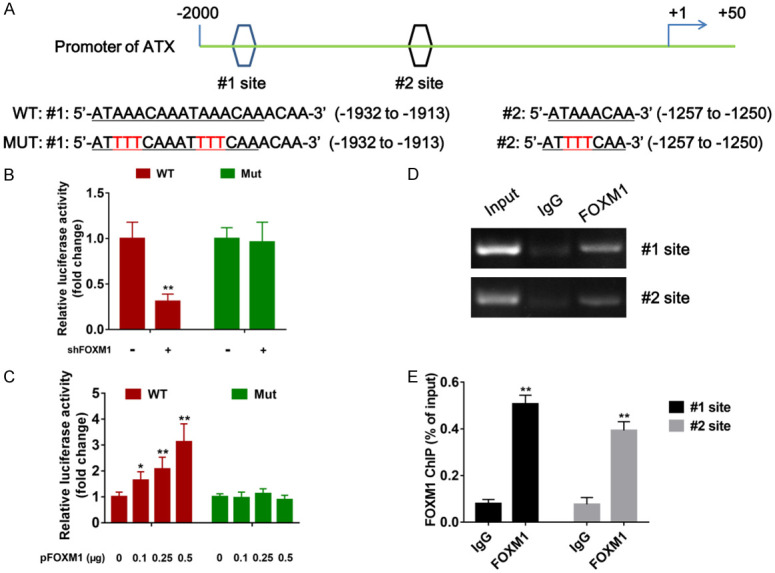
Direct binding of FOXM1 to the ATX promoter. A. Sequences and positions of putative FOXM-binding sites (#1 site and #2 site) in the ATX promoter and two types of ATX promoter (wild type and mutant type) used in dual luciferase reporter assays were shown. B. Stable PANC-1 cells with FOXM1 knockdown were transfected with 0.2 μg of the wild/mutant type of ATX promoter-luciferase construct, and after 24 hours, the promoter activity was examined by dual luciferase reporter assays. C. Normal PANC-1 cells were co-transfected with 0.2 μg of the wild/mutant type of ATX promoter-luciferase construct and 0/0.1/0.25/0.5 μg of FOXM1-expressing plasmids, respectively, and 24 h after transfection, the promoter activity was examined by dual luciferase reporter assays. D. ChIP assays on putative FOXM1-binding sites of the ATX promoter were performed in PANC-1 cells. A specific anti-FOXM1 antibody was used, normal IgG was used as control, and one percent of the total cell lysates was used for PCR before immunoprecipitation (input control). E. ChIP-qPCR analysis of two FOXM1-binding elements on the ATX promoter was conducted using chromatins immunoprecipitated from PANC-1 cells. *P < 0.05, **P < 0.01.
FOXM1 promotes PDAC cell proliferation and migration via ATX
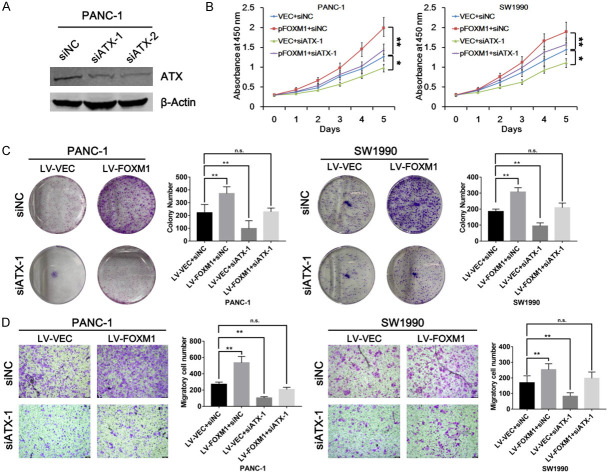
To examine whether FOXM1 exerts its oncogenic function through ATX expression, CCK-8 assays were first employed to test cell growth in PDAC cells. The ATX knockdown was detected in PANC-1 cells by western blot analysis (Figure 3A). Results from CCK-8 assays revealed that ATX knockdown reduced PDAC cell growth compared with control cells. Meanwhile, ATX knockdown impaired the enhanced cell growth induced by FOXM1 overexpression in the two PDAC cell lines: PANC-1 and SW1990 (Figure 3B). Furthermore, we performed colony formation assays using PANC-1 cells and SW1990 cells stably overexpressing FOXM1, and consistently, promotion of colony formation by FOXM1 overexpression was also abolished by ATX knockdown (Figure 3C). On the other hand, cell migration results from transwell assays showed that FOXM1 promoted cell migration dependent on ATX expression in PANC-1 and SW1990 cells (Figure 3D). Therefore, these data above suggested that ATX may mediate the oncogenic role of FOXM1 in PDAC.
Figure 3.
Effects of FOXM1 on PDAC cell proliferation and migration were mediated by regulating ATX expression. A. 10 μL of ATX-specific siRNAs (siATX-1 and siATX-2) were transfected into PANC-1 cells, respectively. And after incubation for 48 h, the efficiencies of siATX-1 and siATX-2 were verified by western blots. B. PANC-1 and SW1990 cells were transfected with pFOXM1 or siATX-1 as indicated, and after 24 h, CCK-8 assays were performed using the indicated PANC-1 and SW1990 cells and cell viability was measured at the indicated time points. C. PANC-1 and SW1990 cells with stable expression of FOXM1 were transfected with siATX-1/siNC as indicated, and colony formation assays were performed 24 h after transfection. D. PANC-1 and SW1990 cells with stable expression of FOXM1 were transfected with siATX-1/siNC as indicated, and cell migration assays were performed 24 h after transfection. *P < 0.05, **P < 0.01.
ATX stimulates FOXM1 expression via inhibiting the Hippo signaling in PDAC cells
Previous researches have clarified that FOXM1 is a downstream target gene of Hippo signaling pathway [21], and inhibition of Hippo pathway will induce dephosphorylation and nuclear localization of its main effector molecule Yes-Associated Protein (YAP), resulting in elevated FOXM1 transcription in tumor cells [22]. Interestingly, recent studies have shown that LPA, a primary downstream target gene of ATX, can suppress the Hippo/YAP signaling and thus mediates the up-regulation of FOXM1 mRNA and protein expression [23,24]. Therefore, we assumed that ATX might regulate FOXM1 expression in PDAC cells through inhibiting the Hippo/YAP signaling. To validate our hypothesis, western blots and qPCR analyses were performed after ATX knockdown in PANC-1 cells. As shown in Figure 4A and 4B, ATX knockdown significantly decreased FOXM1 protein and mRNA expression in PANC-1 cells. Next, we investigated whether this effect could be reversed by XMU-MP-1, a selective Hippo pathway (MST1/2) inhibitor. Western blots analysis suggested that XMU-MP-1 treatment strongly reversed the ATX knockdown-mediated effects on FOXM1 expression (Figure 4C), which was validated by further qPCR analysis (Figure 4D). In addition, a dual luciferase reporter assay was employed to further explore the molecular mechanism via which ATX promoted FOXM1 expression. The results showed that FOXM1 promoter activity was significantly suppressed by knockdown of ATX, which was abolished by the treatment of XMU-MP-1 (Figure 4E), hinting to us that ATX mediated transcriptional activation of FOXM1 by inhibiting the activity of Hippo signaling pathway. These findings mentioned above demonstrated that ATX was critical for FOXM1 expression, and Hippo signaling plays important roles in this process.
Figure 4.
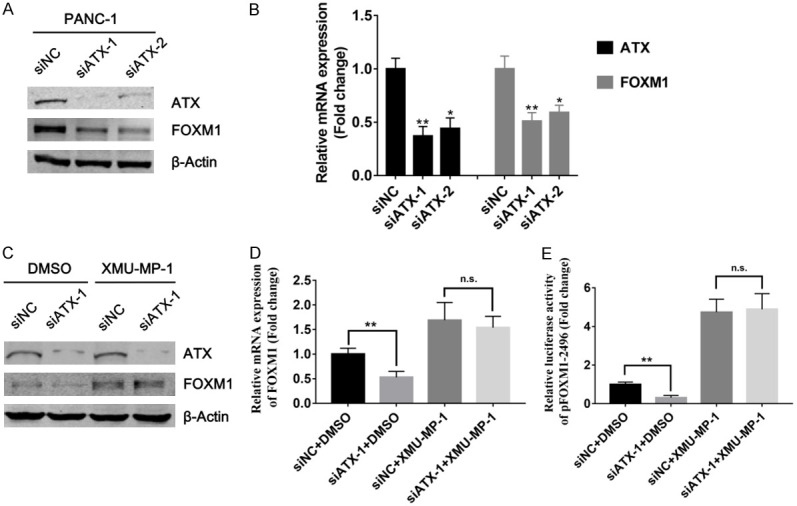
ATX regulated FOXM1 expression via inhibiting the Hippo/YAP signaling. (A and B) 48 h after transfection with siATX-1, proteins or RNAs were isolated from PANC-1 cells and western blots (A) and qPCR (B) analyses were performed to investigate the effects of ATX knockdown on FXOM1 protein and mRNA expression. (C and D) 42 h after transfection with siATX-1, PANC-1 cells were treated with a Hippo signaling-specific inhibitor XMU-MP-1 (3 μM) for 6 h, and western blots (C) and qPCR analyses (D) were performed to examine the expression changes of FOXM1 at protein and mRNA levels. (E) PANC-1 cells were co-transfected with siATX-1/siNC and pFOXM1-2496 for 18 h, and then treated with XMU-MP-1 (3 μM) for 6 h. Finally dual luciferase reporter assays were performed to investigate the changes of FOXM1 promoter activity. *P < 0.05, **P < 0.01.
Positive correlation of elevated expression of ATX and FOXM1 in PDAC
To verify the role of ATX in PDAC development and its relationship with FOXM1, a TMA was used to investigate ATX and FOXM1 protein expression by IHC analysis, and representative images were exhibited in Figure 5A. By statistical analysis, we found that both ATX (Table 1) and FOXM1 (Table 2) expression were significantly higher in tumor tissues as compared with adjacent normal tissues. Moreover, increased FOXM1 expression was positively correlated with elevated ATX expression in tumor tissues (R=0.66, P < 0.01, Figure 5B). These findings further confirmed the oncogenic role of ATX and its positive correlation with FOXM1 in PDAC.
Figure 5.
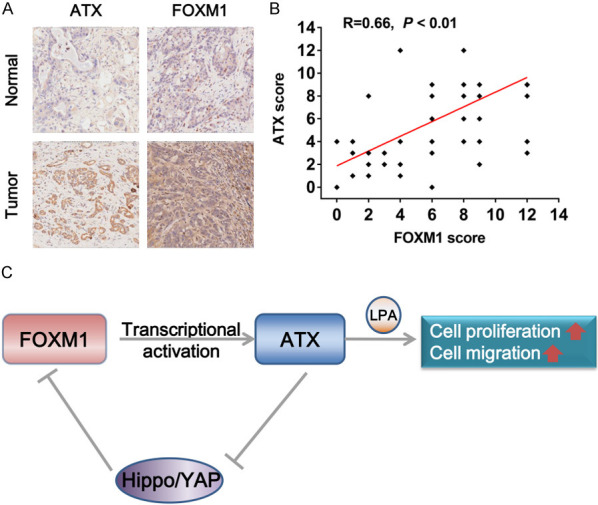
Positive correlation between FOXM1 and ATX expression in PDAC specimens. A. Representative IHC images of FOXM1 and ATX in PDAC tumor tissues and adjacent normal tissues were shown. Magnification: 200 ×. B. The correlation between ATX and FOXM1 expression in PDAC tumor tissues was analyzed using Pearson’s correlation coefficient analysis (n=90, R=0.66, P < 0.01), some of the dots represented more than one specimen. C. A model of the FOXM1/ATX positive feedback loop in PDAC was shown. FOXM1 transcriptionally activated ATX in PDAC cells, and ATX further promoted the transcriptional activity of FOXM1 by inhibiting the Hippo/YAP signaling, thus forming a positive feedback loop, which finally promoted PDAC cell proliferation and migration via inducing the production of its downstream oncogenic genes such as LPA.
Table 1.
Differential expression of ATX between PDAC tumor tissues and adjacent normal tissues
| ATX | Weak (%) | Strong (%) | P value |
|---|---|---|---|
| Normal | 45 (75%) | 15 (25%) | < 0.01 |
| Tumor | 21 (35%) | 39 (65%) |
Table 2.
Differential expression of FOXM1 between PDAC tumor tissues and adjacent normal tissues
| ATX | Weak (%) | Strong (%) | P value |
|---|---|---|---|
| Normal | 37 (62%) | 23 (38%) | < 0.01 |
| Tumor | 18 (30%) | 42 (70%) |
Discussion
In the present study, we demonstrated for the first time that FOXM1 transcriptionally activated ATX, and ATX stimulated FOXM1 expression via inhibiting the Hippo/YAP signaling, thus forming a positive feedback loop and promoted PDAC cell proliferation and migration (Figure 5C).
ATX has been commonly recognized as an oncogene mainly by catalyzing the production of LPA, which enhances cell proliferation, migration and survival in different solid tumors such as prostate, breast, pancreatic and other cancers [25-27]. Previously, several researches have suggested ATX is related with the development of pancreatic cancer. For example, Yatomi et al. found that ATX level in the serum of patients with pancreatic cancer [10], and increased ATX expression was observed in a highly invasive PANC-1 cell line [11]. However, molecular mechanisms underlying aberrant activation of ATX/LPA signaling in cancers remains unclear yet. Herein, we reported that ATX is a downstream target gene of transcription factor FOXM1. Knockdown of FOXM1 significantly increased ATX expression whereas FOXM1 overexpression resulted in reduced ATX expression at both mRNA and protein levels. Furthermore, luciferase reporter assays and ChIP assays demonstrated that FOXM1 bound directly to specific regions of ATX promoter and positively regulated ATX promoter activity. Cell biological experiments suggested that the positive effects of FOXM1 on PDAC cell proliferation and migration abilities were dependent on the regulation of ATX expression. By IHC analysis, we confirmed that both ATX and FOXM1 were frequently up-regulated in PDAC tumor tissues when compared with adjacent normal tissues, and elevated ATX and FOXM1 expression were positively correlated with each other. Therefore, these findings strongly indicated that FOXM1 is a critical regulator of aberrant activation of ATX in PDAC.
As a key regulator of cell proliferation, invasion, metastasis, metabolism, angiogenesis and stem cell maintenance, FOXM1 has been identified as an oncogenic transcription factor in different human malignancies including PDAC [17,18]. However, the molecular mechanisms underlying increased expression of FOXM1 and in PDAC still require further studies. Combined with previous studies that ATX/LPA axis can inhibit the activity of the Hippo signaling pathway and that inhibition of the Hippo signaling promotes the expression and transcriptional activity of FOXM1 through increasing the nuclear localization of YAP [22-24], we assumed that ATX might influence FOXM1 expression in PDAC cells by regulating the Hippo/YAP signaling. By molecular biological techniques we found that knockdown of ATX significantly reduced both mRNA and protein expression of FOXM1 in PDAC cells, which could be greatly reversed by treatment of a specific inhibitor of Hippo signaling pathway (MST1/2), XMU-MP-1. More importantly, results of dual luciferase reporter assays indicated that knockdown of ATX significantly inhibited the activity of FOXM1 promoter, which was strongly attenuated by treatment of XMU-MP-1. Hence, these data identified that transcriptional activation of ATX by FOXM1 increased the transcriptional activity of FOXM1 in turn via inhibiting the Hippo/YAP signaling, thus forming a positive feedback loop and promoted PDAC cell proliferation and migration.
Conclusions
In summary, the present study not only identified a positive feedback loop formed by ATX and FOXM1 and its important role in promoting PDAC cell proliferation and migration, but also indicated that inhibition of both ATX and FOXM1 might be a promising therapeutic strategy against human PDAC.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (No. 81972290 and No. 81602051), Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (No. PWZxq2017-13), Natural Science Foundation of Shanghai (No. 19ZR1441800) and the Outstanding Clinical Discipline Project of Shanghai Pudong (PWYgy2018-02).
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 4.Tokumura A, Majima E, Kariya Y, Tominaga K, Kogure K, Yasuda K, Fukuzawa K. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J Biol Chem. 2002;277:39436–39442. doi: 10.1074/jbc.M205623200. [DOI] [PubMed] [Google Scholar]
- 5.van Meeteren LA, Moolenaar WH. Regulation and biological activities of the autotaxin-LPA axis. Prog Lipid Res. 2007;46:145–160. doi: 10.1016/j.plipres.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Umezu-Goto M, Kishi Y, Taira A, Hama K, Dohmae N, Takio K, Yamori T, Mills GB, Inoue K, Aoki J, Arai H. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J Cell Biol. 2002;158:227–233. doi: 10.1083/jcb.200204026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 8.Federico L, Jeong KJ, Vellano CP, Mills GB. Autotaxin, a lysophospholipase D with pleomorphic effects in oncogenesis and cancer progression. J Lipid Res. 2016;57:25–35. doi: 10.1194/jlr.R060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee D, Suh DS, Lee SC, Tigyi GJ, Kim JH. Role of autotaxin in cancer stem cells. Cancer Metastasis Rev. 2018;37:509–518. doi: 10.1007/s10555-018-9745-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakai Y, Ikeda H, Nakamura K, Kume Y, Fujishiro M, Sasahira N, Hirano K, Isayama H, Tada M, Kawabe T, Komatsu Y, Omata M, Aoki J, Koike K, Yatomi Y. Specific increase in serum autotaxin activity in patients with pancreatic cancer. Clin Biochem. 2011;44:576–581. doi: 10.1016/j.clinbiochem.2011.03.128. [DOI] [PubMed] [Google Scholar]
- 11.Fukushima K, Otagaki S, Takahashi K, Minami K, Ishimoto K, Fukushima N, Honoki K, Tsujiuchi T. Promotion of cell-invasive activity through the induction of LPA receptor-1 in pancreatic cancer cells. J Recept Signal Transduct Res. 2018;38:367–371. doi: 10.1080/10799893.2018.1531889. [DOI] [PubMed] [Google Scholar]
- 12.Bella L, Zona S, Nestal de Moraes G, Lam EW. FOXM1: a key oncofoetal transcription factor in health and disease. Semin Cancer Biol. 2014;29:32–39. doi: 10.1016/j.semcancer.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13:482–495. doi: 10.1038/nrc3539. [DOI] [PubMed] [Google Scholar]
- 14.Xie Z, Tan G, Ding M, Dong D, Chen T, Meng X, Huang X, Tan Y. Foxm1 transcription factor is required for maintenance of pluripotency of P19 embryonal carcinoma cells. Nucleic Acids Res. 2010;38:8027–8038. doi: 10.1093/nar/gkq715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 16.Wierstra I. The transcription factor FOXM1 (Forkhead box M1): proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles. Adv Cancer Res. 2013;118:97–398. doi: 10.1016/B978-0-12-407173-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 17.Wierstra I. FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv Cancer Res. 2013;119:191–419. doi: 10.1016/B978-0-12-407190-2.00016-2. [DOI] [PubMed] [Google Scholar]
- 18.Quan M, Wang P, Cui J, Gao Y, Xie K. The roles of FOXM1 in pancreatic stem cells and carcinogenesis. Mol Cancer. 2013;12:159. doi: 10.1186/1476-4598-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nandi D, Cheema PS, Jaiswal N, Nag A. FoxM1: repurposing an oncogene as a biomarker. Semin Cancer Biol. 2018;52:74–84. doi: 10.1016/j.semcancer.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Li Z, Jia Z, Gao Y, Xie D, Wei D, Cui J, Mishra L, Huang S, Zhang Y, Xie K. Activation of vitamin D receptor signaling downregulates the expression of nuclear FOXM1 protein and suppresses pancreatic cancer cell stemness. Clin Cancer Res. 2015;21:844–853. doi: 10.1158/1078-0432.CCR-14-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisinger-Mathason TS, Mucaj V, Biju KM, Nakazawa MS, Gohil M, Cash TP, Yoon SS, Skuli N, Park KM, Gerecht S, Simon MC. Deregulation of the Hippo pathway in soft-tissue sarcoma promotes FOXM1 expression and tumorigenesis. Proc Natl Acad Sci U S A. 2015;112:E3402–11. doi: 10.1073/pnas.1420005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizuno T, Murakami H, Fujii M, Ishiguro F, Tanaka I, Kondo Y, Akatsuka S, Toyokuni S, Yokoi K, Osada H, Sekido Y. YAP induces malignant mesothelioma cell proliferation by upregulating transcription of cell cycle-promoting genes. Oncogene. 2012;31:5117–5122. doi: 10.1038/onc.2012.5. [DOI] [PubMed] [Google Scholar]
- 23.Fan Q, Cai Q, Xu Y. FOXM1 is a downstream target of LPA and YAP oncogenic signaling pathways in high grade serous ovarian cancer. Oncotarget. 2015;6:27688–27699. doi: 10.18632/oncotarget.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, Fu XD, Mills GB, Guan KL. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valdes-Rives SA, Gonzalez-Arenas A. Autotaxin-lysophosphatidic acid: from inflammation to cancer development. Mediators Inflamm. 2017;2017:9173090. doi: 10.1155/2017/9173090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leblanc R, Peyruchaud O. New insights into the autotaxin/LPA axis in cancer development and metastasis. Exp Cell Res. 2015;333:183–189. doi: 10.1016/j.yexcr.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Lv GM, Li P, Wang WD, Wang ShK, Chen JF, Gong YL. Lysophosphatidic acid (LPA) and endothelial differentiation gene (Edg) receptors in human pancreatic cancer. J Surg Oncol. 2011;104:685–691. doi: 10.1002/jso.22016. [DOI] [PubMed] [Google Scholar]