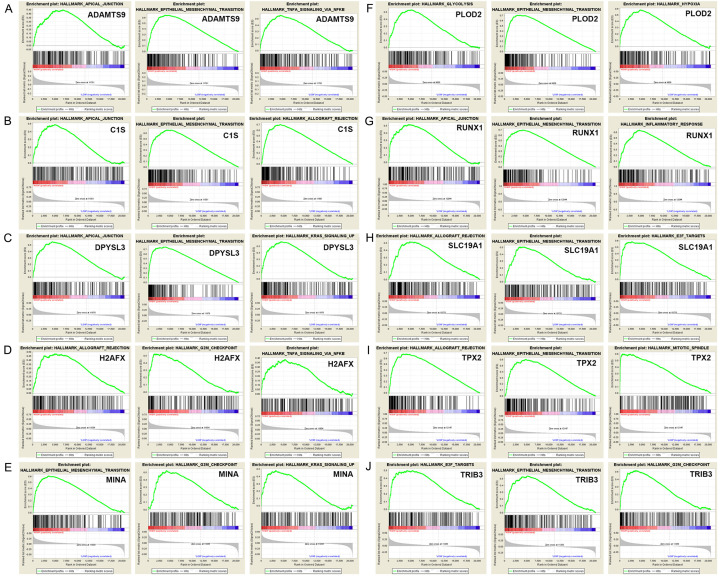
Figure 8.
GSEA was used to perform hallmark analysis for ADAMTS9, C1S, DPYSL3, H2AFX, MINA, PLOD2, RUNX1, SLC19A1, TPX2 and TRIB3. GSEA identified 100 DEGs with positive or negative correlations. Among them, we used the hallmarks ADAMTS9, C1S, DPYSL3, H2AFX, MINA, PLOD2, RUNX1, SLC19A1, TPX2, and TRIB3 to perform GSEA. Results suggested that the most involved three significant pathways of each oncogene included apical junction, epithelial mesenchymal transition, allograft rejection, G2M checkpoint, mitotic spindle, Kras signaling pathway up, inflammatory response, TNF-alpha signaling via NF-κB, etc.