Abstract
Paenibacillus curdlanolyticus B-6 is a facultative anaerobic bacterium that efficiently produces a lignocellulolytic multienzyme complex. The whole genome of P. curdlanolyticus B-6 was sequenced on an Ion GeneStudio S5 system, which yielded 74 contigs with a total size of 4,875,097 bp, 4,473 protein-coding sequences, and a G+C content of 49.7%. The genome data have been deposited in DDBJ/ENA/GenBank under accession numbers BLWM01000001–BLWM01000074. Analyses of average nucleotide identities and phylogenetic relationships of 16S rRNA sequences of Paenibacillus species revealed that strain B-6 is most closely related to Paenibacillus xylaniclasticus TW1. P. curdlanolyticus B-6 should thus be reclassified as a strain of P. xylaniclasticus.
Keywords: Paenibacillus curdlanolyticus B-6, Draft genome, Xylanolytic enzyme, Cellulolytic enzyme, Multienzyme complex
Specifications Table
| Subject | Microbiology |
| Specific subject area | Bacteriology, Genomics |
| Type of data | Table, Figures |
| How data were acquired | Whole-genome sequencing using Ion GeneStudio S5 System |
| Data format | Raw and Analyzed |
| Parameters for data collection | Genomic DNA was extracted from pure culture of P. curdlanolyticus B-6. The genome of strain B-6 was sequenced by using Ion GeneStudio S5 System, de novo assembled using CLC Genomic Workbench 20.0.1, and annotated using DDBJ Fast Annotation and Submission Tool (DFAST). |
| Description of data collection | Genomic DNA extracted from P. curdlanolyticus B-6, following whole-genome sequencing, assembly, and annotation |
| Data source location | Japan International Research Center for Agricultural Sciences (JIRCAS) Tsukuba, Ibaraki, Japan |
| Data accessibility | Repository name: DDBJ/ENA/GenBank Data identification number: BLWM01000000. The version described in this paper is BLWM01000000.1 Direct URL to data: https://www.ncbi.nlm.nih.gov/nuccore/BLWM00000000.1 The BioProject ID in GenBank is PRJDB9861 (https://www.ncbi.nlm.nih.gov/bioproject/PRJDB9861) The BioSample ID in GenBank is SAMD00228050 (https://www.ncbi.nlm.nih.gov/biosample/?term=SAMD00228050) |
| Related research article | P. Pason, K-L. Kyu, K. Ratanakhanokchai, Paenibacillus curdlanolyticus strain B-6 xylanolytic-cellulolytic enzyme system that degrades insoluble polysaccharides, Appl. Environ. Microbiol. 72 (2006) 2483–2490. 10.1128/AEM.72.4.2483–2490.2006 |
Value of the Data
-
•
Paenibacillus curdlanolyticus B-6 produces a large extracellular complex enzyme, which is unusual in cellulolytic-xylanolytic Paenibacillus species.
-
•
Genome data of strain B-6 will be useful for further functional genomics and enzyme engineering research.
-
•
The draft genome sequence of strain B-6 can aid understanding of the polysaccharide degradation mechanism of this bacterium and may be useful as a reference sequence for Paenibacillus species classification.
1. Data Description
Bacterial enzyme systems for lignocellulose degradation can be generally regarded as non-complexed or complexed enzymes that are normally produced by aerobic and anaerobic bacteria, respectively. In terms of hydrolysis efficiency, the complexed enzymes offer greater potential for the degradation of lignocellulose compared with non-complexed ones. The production of enzymes by anaerobic culture is very costly, however, mainly because of the high price of medium, slow rate of growth, and low enzyme yield [1].
The mesophilic facultatively anaerobic bacterium Paenibacillus curdlanolyticus strain B-6, isolated from an anaerobic digester fed with pineapple wastes [2], was originally classified according to the results of a 16S rRNA gene analysis by Pason et al. [3]. Strain B-6 is a true lignocellulolytic microorganism, as it can use xylan, microcrystalline cellulose, and lignocellulosic biomass as sole carbon sources [3]. Strain B-6 was found to produce complexed enzymes under aerobic conditions [4, 5], a rarely reported phenomenon [6,7,8,9]. In recent years, the characteristics and function of the lignocellulolytic enzyme system of this bacterium have been the subject of considerable research. We found that the complex enzyme produced by strain B-6 is critical for improving lignocellulosic biomass degradation; however, the mechanisms of lignocellulose degradation and utilization are still unclear. A similar bacterial example, Paenibacillus xylaniclasticus strain TW1 [10] isolated from sludge in an anaerobic digester, is known to have a xylan degradation system, as in strain B-6. Because of differences in several phenotypic characters, such as growth temperature and acid formation [10], we have not previously analyzed the taxonomic relationship of strains B-6 and TW1. An understanding of the genetic relationship of the two strains and differences in their xylan degradation systems was thus needed.
In this work, we determined the draft genome sequence of strain B-6 to obtain further information on lignocellulose utilization systems in the genus Paenibacillus. Features of the genome are shown in Table 1. DNA sequencing, performed using the Ion GeneStudio S5 System, generated 45,085,168 reads. The genome was assembled de novo using CLC Genomic Workbench 20.0.1 (CLC Bio, Qiagen, Valencia, CA), which resulted in 74 contigs with an N50 of 237,553 bp and a maximum size of 430,139 bp. The genome of P. curdlanolyticus strain B-6 comprised 4,875,097 bp and had a G + C content of 49.7%, which is nearly identical to that of P. xylaniclasticus (4,924,585 bp, with a G + C content of 49.6%). Genome annotation was performed with the DDBJ Fast Annotation and Submission Tool (DFAST). Paenibacillus curdlanolyticus strain B-6 was found to have 4,473 protein-coding sequences (CDSs), 4 rRNA genes, and 94 tRNA genes.
Table 1.
Features of the Paenibacillus curdlanolyticus B-6 genome.
| Feature | Description |
|---|---|
| Number of reads used in the assembly | 45,085,168 |
| Mean read length | 195 bp |
| Genome size | 4,875,097 bp |
| Number of contigs | 74 |
| G + C content (%) | 49.7 |
| N50 contig length | 237,553 bp |
| Mean contig length | 65,880 bp |
| Number of CDSs | 4,473 |
| Number of rRNAs | 4 |
| Number of tRNAs | 94 |
| Number of CRISPRs | 5 |
| Genome coverage depth | 1,718-fold |
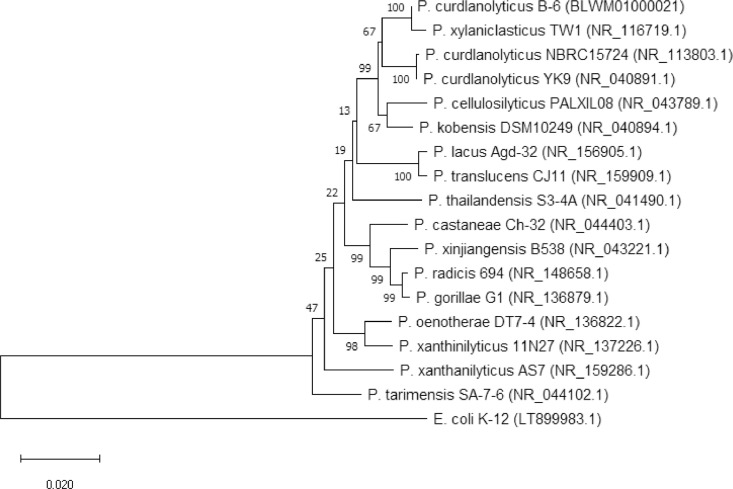
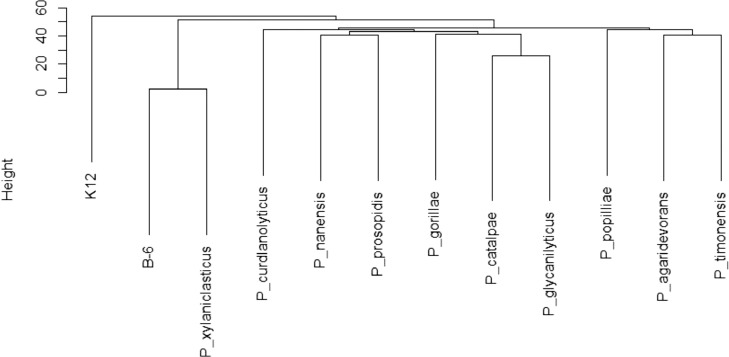
Phylogenetic analysis of 16S rRNA sequences of strain B-6 and 16 related strains revealed that strain B-6 is very closely related to P. xylaniclasticus TW1 (99.51% similarity), P. curdlanolyticus NBRC15724 (97.74% similarity), and P. curdlanolyticus YK9 (97.67% similarity) (Fig. 1 and Suppl. Table 1). Moreover, an assay of the average nucleotide identity (ANI) [11] of 11 strains belonging in 10 Paenibacillus species, including strain B-6, and one outgroup strain, Escherichia coli K-12 (LT899983), showed that strain B-6 is more closely related to P. xylaniclasticus TW1 than to P. curdlanolyticus YK9 (Fig. 2, Suppl. Table 2 and 3).
Fig. 1.
A 16S rRNA-based phylogenetic tree of Paenibacillus curdlanolyticus B-6 and related members of the genus Paenibacillus. Escherichia coli K-12 was used as an outgroup. The phylogenetic tree was constructed using the neighbor-joining method with 1,000 bootstrap replicates. The bar represents 0.02 substitutions per nucleotide position.
Fig. 2.
Dendrogram of average nucleotide identity (ANI) values. The ANI value of each strain was calculated, and a dendrogram was constructed using the unweighted pair group method with arithmetic means. Escherichia coli K-12 (NZ_CP014272) was used as an outgroup. Strains of Paenibacillus were as follows: Paenibacillus agaridevorans (NZ_BDQX01000000), P. catalpae (NZ_FOMT01000000), P. curdlanolyticus (NZ_AEDD01000000), P. glycanilyticus (NZ_BILY01000000), P. gorillae (NZ_CBVJ010000000), P. nanensis (NZ_QXQA01000000), P. popilliae (NZ_BALG01000000), P. prosopidis (NZ_QPJD01000000), P. timonensis (NZ_WNZY01000000), and P. xylaniclasticus (NZ_BIML01000000).
Although analysis of the genome data of strain B-6 demonstrated its high similarity to strain TW1, a previous investigation of enzyme component patterns of both strains clearly indicated they have different xylanase profiles [3, 10]. In addition, BLAST searching with the B-6 draft sequence as the query failed to uncover two characteristic xylanases of strain B-6, namely, Xyn10D [12] and Xyn10E [13], in the P. xylaniclasticus TW1 genome. We therefore believe that taxonomic analysis of strains B-6 and TW1 is necessary.
2. Experimental design, materials and methods
2.1. Genomic DNA extraction and sequencing
Genomic DNA of P. curdlanolyticus B-6 was obtained by phenol/chloroform extraction from cells grown under aerobic conditions at 37 °C. Fragmentation of DNA was performed with a Bioruptor sonicator (BMBio, Japan), which generated fragments with an average length of 500 bp. Approximately 400- to 600-bp fragments were size-selected by electrophoresis on E-Gel SizeSelect II agarose gels (Invitrogen, Thermo Fisher Scientific) before library preparation. The DNA library was prepared using an Ion Plus Fragment Library kit (Thermo Fisher Scientific) according to the manufacturer's protocol. The genomic DNA of P. curdlanolyticus B-6 was sequenced using an Ion GeneStudio S5 System.
2.2. Phylogenetic species identification
The 16S rRNA sequence of strain B-6 was analyzed using the BLAST search engine and manually aligned with sequences in the GenBank database using the Multiple Sequence Alignment option in CLUSTAL W (https://www.genome.jp/tools-bin/clustalw). Phylogenetic trees were constructed by the neighbor-joining method using MEGA version 10.1.8 software [14]. Tree topologies and distances were estimated by performing a bootstrap analysis with 1,000 re-samplings.
2.3. Genome assembly and annotation
After removal of low-quality reads, de novo genome assembly was performed using CLC Genomic Workbench version 20.0.1. The genome was annotated using DFAST (https://dfast.nig.ac.jp/). An additional analysis was performed using the carbohydrate-active enzymes (CAZy) database (http://www.cazy.org/).
2.4. Genomic ANI
Pairwise ANI values of whole genome sequences of Paenibacillus strains were calculated using GENETYX NGS version 4.1.1. The matrix generated from ANI values among Paenibacillus strains was converted to a genetic dendrogram using algorithms such as the unweighted pair group method with arithmetic means and the single-linkage clustering method in the R statistical program.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgments
This work was conducted as part of a development project funded by Exploratory Research for Advanced Technology (ERATO) (grant number JPMJER1502) of the Japan Science and Technology Agency (JST) and the Science and Technology Research Partnership for Sustainable Development (SATREPS) (grant number JPMJSA1801) of the Japan Science and Technology Agency (JST)/Japan International Cooperation Agency (JICA). We thank Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.106213.
Appendix. Supplementary materials
References
- 1.Ratanakhanokchai K., Waeonukul R., Pason P., Tachaapaikoon C., Kyu K.L., Sakka K., Kosugi A., Mori Y. Biomass Now-Cultivation and Utilization. IntechOpen; 2013. Paenibacillus curdlanolyticus strain B-6 multienzyme complex: a novel system for biomass utilization; pp. 369–394. [DOI] [Google Scholar]
- 2.M. Tanticharoen, S. Cheevadhanarak, The production of cellulase and xylanase from cellulolytic microorganisms isolated from pineapple anaerobic digester. II. Activities Studies. Annual report of ASEAN Working Group on the Management and Utilization of Food Waste Materials (1984) 493–505.
- 3.Pason P., Kyu K.L., Ratanakhanokchai K. Paenibacillus curdlanolyticus strain B-6 xylanolytic-cellulolytic enzyme system that degrades insoluble polysaccharides. Appl. Environ. Microbiol. 2006;72:2483–2490. doi: 10.1128/AEM.72.4.2483-2490.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waeonukul R., Kyu K.L., Sakka K., Ratanakhanokchai K. Isolation and characterization of a multienzyme complex (cellulosome) of the Paenibacillus curdlanolyticus B-6 grown on Avicel under aerobic conditions. J. Biosci. Bioeng. 2009;107:610–614. doi: 10.1016/j.jbiosc.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Pason P., Kosugi A., Waeonukul R., Tachaapaikoon C., Ratanakhanokchai K., Arai T., Murata Y., Nakajima J., Mori Y. Purification and characterization of a multienzyme complex produced by Paenibacillus curdlanolyticus B-6. Appl. Microbiol. Biotechnol. 2010;85:573–580. doi: 10.1007/s00253-009-2117-2. [DOI] [PubMed] [Google Scholar]
- 6.Kim C.H., Kim D.S. Extracellular cellulolytic enzymes of Bacillus circulans are present as two multipleprotein complexes. Appl. Biochem. Biotechnol. 1993;42:83–94. doi: 10.1007/BF02788904. [DOI] [Google Scholar]
- 7.Hou P.B., Li Y.Z., Wu B.H., Yan Z.C., Yan B.X., Gao P.J. Cellulolytic complex exists in cellulolytic myxobacterium Sorangium. Enzyme Microb. Technol. 2006;38:273–278. doi: 10.1016/j.enzmictec.2004.08.044. [DOI] [Google Scholar]
- 8.van Dyk J.S., Sakka M., Sakka K., Pletschke B.I. The cellulolytic and hemi-cellulolytic system of Bacillus licheniformis SVD1 and the evidence for production of a large multi-enzyme complex. Enzyme Microb. Technol. 2009;45:372–378. doi: 10.1016/j.enzmictec.2009.06.016. [DOI] [Google Scholar]
- 9.Jones S.M., van Dyk J.S., Pletschke B.I. Bacillus subtilis SJ01 produces hemicellulose degrading multi-enzyme complexes. BioResources. 2012;7:1294–1309. [Google Scholar]
- 10.Tachaapaikoon C., Tanasupawat S., Pason P., Sornyotha S., Waeonukul R., Kyu K.L., Ratanakhanokchai K. Paenibacillus xylaniclasticus sp. nov., a xylanolytic-cellulolytic bacterium isolated from sludge in an anaerobic digester. J. Microbiol. 2012;50:394–400. doi: 10.1007/s12275-012-1480-3. [DOI] [PubMed] [Google Scholar]
- 11.Goris J., Konstantidis K.T., Klappenbach J.A., Coenye T., Vandamme P., Tiedje J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 12.Sakka M., Higashi Y., Kimura T., Ratanakhanokchai K., Sakka K. Characterization of Paenibacillus curdlanolyticus B-6 Xyn10D, a xylanase that contains a family 3 carbohydrate-binding module. Appl. Environ. Microbiol. 2011;77:4260–4263. doi: 10.1128/AEM.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sermsathanaswadi J., Baramee S., Tachaapaikoon C., Pason P., Ratanakhanokchai K., Kosugi A. The family 22 carbohydrate-binding module of bifunctional xylanase/β-glucanase Xyn10E from Paenibacillus curdlanolyticus B-6 has an important role in lignocellulose degradation. Enzyme Microb. Technol. 2017;96:75–84. doi: 10.1016/j.enzmictec.2016.09.015. [DOI] [PubMed] [Google Scholar]
- 14.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.