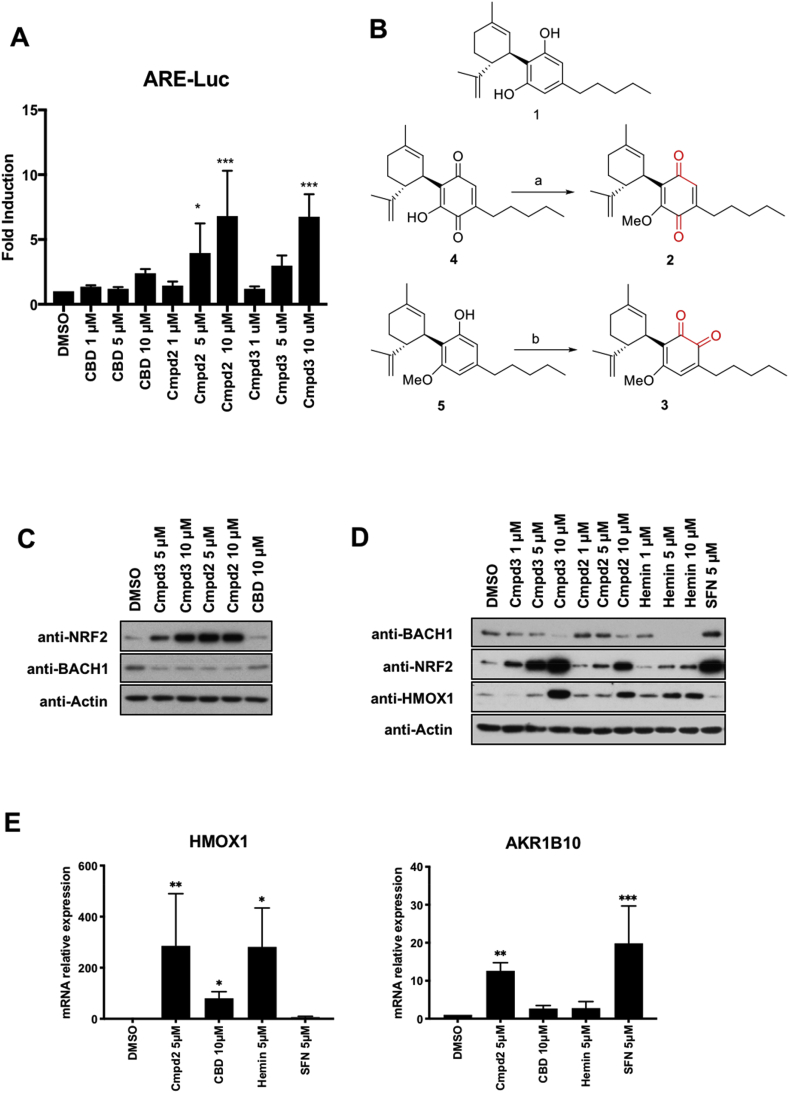
Fig. 1.
Screening of CBD derivatives and characterization of compound 2 as a BACH1 inhibitor and NRF2 inducer. A) HaCaT-ARE-Luc cells were treated with either DMSO or increasing concentrations of CBD, compound 2 (Cmpd2) or compound 3 (Cmpd3) for 6 h. Luciferase activity was measured in the cell lysates and expressed as RLU (x 104). Data represent means ± SD (n = 4) and are expressed relative to untreated cells. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. B) Schematic representation of the sinthesis of the isomeric methoxyquinones 2 (compound 2) and 3 (compound 3) from 1 (CBD). (a): MeI, NaHCO3, DMF, 49%; (b) SIBX, EtOAc, 51%. C) HaCaT cells were incubated with either DMSO, compound 3 (Cmpd3), compound 2 (Cmpd2) or CBD. Three hours later, cells were lysed and samples were analysed by Western Blot. D) HaCaT cells were incubated with either DMSO, or increasing concentrations of compound 3 (Cmpd3), compound 2 (Cmpd2), hemin or sulforaphane (SFN). Three hours later, cells were lysed and levels of BACH1, NRF2, HMOX1 and Actin were analysed by Western Blot. A representative blot is shown; the corresponding quantifications of BACH1, NRF2 and HMOX1 protein levels are shown in Suppl. Fig. S1A. E) HaCaT cells were treated with either DMSO, compound 2 (5 μM), CBD (10 μM), Hemin (5 μM) or SFN (5 μM) for 8 h. The mRNA levels of HMOX1 and AKR1B10 were quantified by real-time PCR and the data were normalised using HPRT1 as an internal control. Data represent means ± SD (n = 3) and are expressed relative to the DMSO sample. Statistical analysis was performed against the DMSO sample. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.