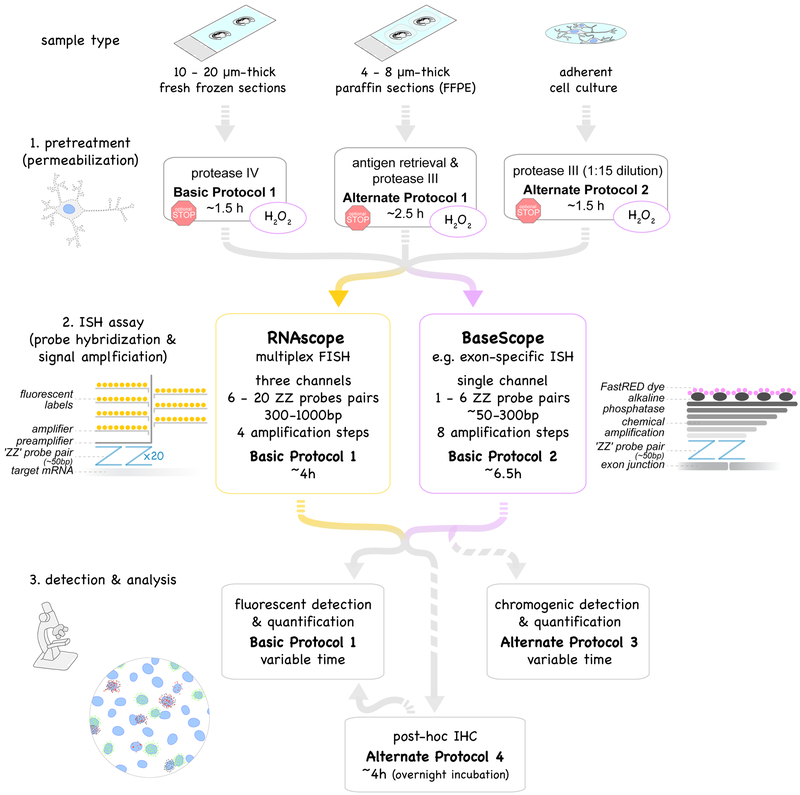
Figure 1.
Overview of protocols described in this unit. Multiplex fluorescent in situ hybridization, RNAscope (yellow box, Basic Protocol 1) and exon-specific in situ hybridization, BaseScope (purple box, Basic Protocol 2) use probe pairs (called “ZZ” pairs) each targeting about 50 bases of target mRNA. While RNAscope uses 6 - 20 ZZ pairs (targeting about 300 - 1000 bases), improved signal amplification allows BaseScope assays to target short RNA sequences (50 - 300 bases) by using only 1 - 6 ZZ probe pairs. Signal amplification of both assays are schematically represented and involve 4 and 8 amplification steps, respectively. Fluorescent labels in RNAscope are detected using a fluorescent microscope (Basic Protocol 1). The FastRED dye used in BaseScope and converted by alkaline phosphatase is both visible under fluorescent and light microscope (Basic Protocol 2). Both RNAscope and BaseScope are compatible with different sample types: 10-20μm-thick fresh-frozen sections (Basic Protocol 1), 4–8μm-thick formalin-fixed paraffin-embedded (FFPE) sections (Alternate Protocol 1) and adherent cell cultures such as neuronal cultures (Alternate Protocol 2). Pretreatment and permeabilization to allow the probes to perfuse into the tissue to the target RNA need to be adjusted accordingly. BaseScope assays require an additional pretreatment step with H2O2 to saturate endogenous peroxidase activity (purple circles). Protocols can be interrupted at optional stopping points during pretreatments (red stop sign). Post-assay immunostaining (Basic Protocol 3) is optional.