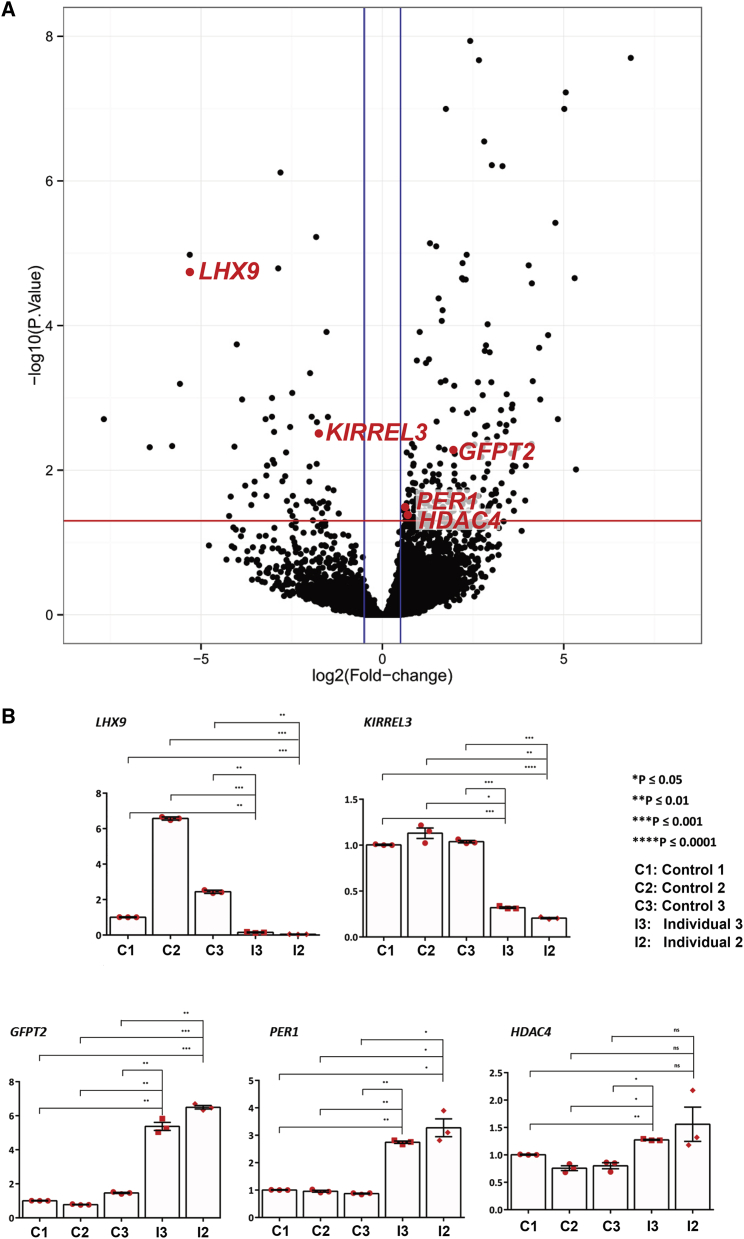
Figure 3.
RNaseq Was Performed on Fibroblasts from Individuals 2 and 3 and Six Healthy Controls
(A) Volcano plot showing common DEGs (differentially expressed genes) of individuals 2 and 3. Significant DEGs. The red line indicates a −log10 (adjusted p value) of 1.3 (padj of 0.05); and the blue line a Log2 Fold Change of −0.05 and 0.05. Significant DEGs shown in panel B are represented by red dots.
(B) Reverse transcriptase-qPCR analysis of specific genes deregulated in fibroblasts from three new controls and from individuals 2 and 3. β-actin was used as the reference gene. Triplicates were used. Error bars represent standard deviation. p values were generated through the use of two-way ANOVA with Dunnett’s multiple comparisons test.