Figure 5.
Location and Functional Impact of Pathogenic Variants
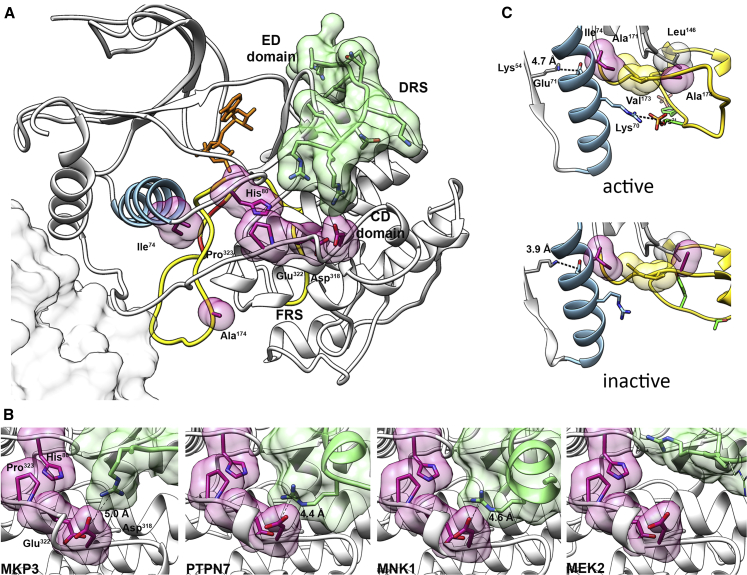
(A) Structure of MAPK1 complexed with a peptide from the D-motif of MKP3 (PDB: 2FYS). Different colors and representations highlight functional important regions. Activation segment, yellow; phosphorylatable residues Thr185 and Tyr187, red; helix C, light blue; mutated residues, semi-transparent violet surfaces; MKP3 D-motif, semi-transparent green surface; putative position of the second monomer in the MAPK1 dimer (according to PDB: 2ERK), gray surface. CD and ED domains, part of the DRS, are indicated. The ATP analog phosphoaminophosphonic acid-adenylate ester, not present in the structure, has been added in the corresponding position from the MAPK1/MNK1 complex (PDB: 2Y9Q) (orange sticks).
(B) Structures of MAPK1 complexes with peptides interacting with the DRS region, corresponding to D-motifs of phosphatases MKP3 (PDB: 2FYS) and PTPN7 (PDB: 2GPH), MAPK1 substrate MNK1 (PDB: 2Y9Q), and kinase MEK2 (PDB: 4H3Q). Representations and colors are used as above. The residues of the interacting peptides forming salt bridges with Asp318 are reported as sticks. In the case of MEK2, no residues form an ion pair with Asp318: the MEK2 residue closest to Asp318 is Arg4, whose side chain was not solved in the crystal structure, indicating that a stable salt bridge between MAPK1-Asp318 and MEK2-Arg4 residues is not present. A putative conformation for Arg4 has been modeled in the structure (thin sticks).
(C) Location and interactions of residues Ile74 and Ala174 in the active and inactive structures of MAPK1 (PDB: 2ERK and 1ERK, respectively). Representations and colors are used as above. Ile74, located on helix C, participates in a hydrophobic cluster comprising activation segment residues Ala171 and Val173, in both protein conformations. This region is critical in the activation mechanism. In the active state, Lys70 forms an ion pair with the phosphorylated Thr185, causing a remodeling of helix C, and the formation of a stronger salt bridge between conserved residues Glu71 and β3 Lys54, which is a prerequisite for activation. Ala174 is in the activation segment and interacts with Leu146, which is in the catalytic region. In all cases, residue numbers correspond to the human sequence.