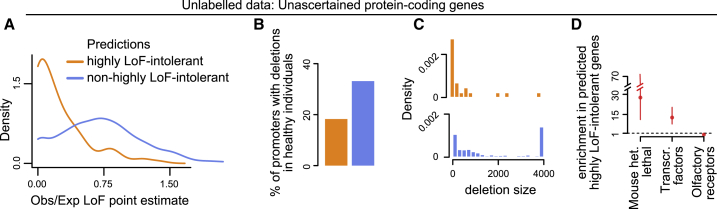
Figure 5.
Using predLoF-CpG to Classify Currently Unascertained Genes as Highly Loss-of-Function Intolerant or Not
(A) The distribution of point estimates of the observed/expected proportions of LoF variants. Genes are stratified according to their classification as highly LoF intolerant or not.
(B) The proportion of promoters which harbor deletions in a sample of 14,891 healthy individuals. Promoters are stratified according to downstream gene classification as highly LoF intolerant or not.
(C) The distribution of the size of deletions harbored by promoters in a sample of 14,891 healthy individuals. Promoters are stratified according to downstream gene classification as highly LoF intolerant or not.
(D) Odds ratios and the corresponding 95% confidence intervals quantifying the enrichment for genes in each of the x axis groups that is exhibited by genes predicted as highly LoF intolerant by predLoF-CpG. The enrichment is computed against genes predicted as non-highly LoF intolerant. The horizontal line at 1 corresponds to zero enrichment.