Abstract
Purpose: We recently reported that tripartite motif-containing 67 (TRIM67) activates p53 to suppress colorectal cancer (CRC). However, the function and mechanism of TRIM67 in the inhibition of CRC cell proliferation and metastasis remains to be further elucidated.
Methods: We detected the expression of TRIM67 in CRC tissues compared with normal tissues and confirmed its relationship with clinicopathological features. DNA methylation of TRIM67 was analyzed to determine its significantly hypermethylated sites in CRC tissues. CCK-8, colony formation, transwell migration, and Matrigel invasion assays were performed to evaluate the effects of TRIM67 on cell proliferation and metastasis in CRC cells. RNA sequencing of TRIM67 and TRIM67 rescue experiments were performed to reveal its mechanisms in CRC cell proliferation and metastasis.
Results: TRIM67 expression was significantly downregulated in CRC tissues and its expression was associated with clinical stage, invasive depth, tumor size, lymph node metastasis, and Dukes' stage. Three methylation sites were significantly hypermethylated and negatively correlated with TRIM67 expression in CRC tissues. TRIM67 suppressed proliferation, migration, and invasion in CRC cells. RNA sequencing revealed that protein mitogen-activated protein kinase 11 (MAPK11) was a potential downstream negative regulatory gene of TRIM67. Reversing MAPK11 expression could rescue the effects of TRIM67 on the proliferation and metastasis of CRC cells.
Conclusion: TRIM67 inhibited cell proliferation and metastasis by mediating MAPK11 in CRC, and may be a potential target to inhibit CRC metastasis.
Keywords: TRIM67, colorectal cancer, MAPK11, cell proliferation, metastasis
Introduction
Colorectal cancer (CRC) is a main cause of death worldwide, and its morbidity and mortality have increased in recent years 1. Multiple genes and signaling pathways have been identified but the precise molecular mechanism of CRC is not fully understood. Therefore, clarifying its pathogenesis and identifying novel molecular biomarkers to improve the diagnosis and prognosis of patients with CRC are urgently required 2.
The tripartite motif (TRIM) family is a large class of RBCC proteins of which nearly 70 members have been identified in humans, while fewer than 20 members have been identified in invertebrates and none in single-celled organisms. The high number of TRIM members in humans indicates that it is a rapidly evolving family 3. Members of the TRIM protein family are involved in many biological processes including proliferation, differentiation, development, autophagy, and apoptosis of cancer cells 4-8. In recent years, various studies have associated several TRIM family members, TRIM15, TRIM27 and TRIM29, with CRC through AKT or JAK2/STAT3 signaling to regulate CRC cell migration and epithelial-mesenchymal transition 9-11.
TRIM67, a TRIM family member, is a protein-coding gene located at 1q42.2 consisting of approximately 59 kb DNA bases that encodes TRIM67 (84 kDa) 12. To our knowledge, TRIM67 has been reported to be involved in neuroprotective effects and tumor processes. Nicholas P. Boyer et al. 13 found that TRIM67 is necessary for appropriate brain development and behavior. TRIM67 is also associated with ubiquitination and interacts with PRG-1 and 80K-H to regulate the RAS signaling pathway to inhibit cell proliferation and enhance synapse production in neuroblastoma 14. Le Duy Do et al. 15 found that autoantibodies against TRIM67 appeared to be specific biomarkers of paraneoplastic cerebellar degeneration and associated with lung carcinoma. A recent study implicated that TRIM67 activates the NF-κB pathway and promotes cell apoptosis in GA-13315-treated lung cancer cells 16. Our previous study found that in The Cancer Genome Atlas (TCGA), TRIM67 is frequently methylated in CRC compared with adjacent normal tissues, and prevents intestinal tumorigenesis in mice and improves chemotherapy efficacy by activating the TRIM67-p53 axis 17. In the present study, TRIM67 expression levels and clinicopathological features based on TCGA dataset and our cohort was further examined. We also investigated the direct functions of TRIM67 in CRC cells and studied its potential mechanisms as a tumor suppressor in CRC.
Materials and Methods
TCGA data download and analysis
mRNA sequencing data of 647 CRC tissues and 51 normal tissues was downloaded from TCGA (https://tcga-data.nci.nih.gov/), and data for the mRNA expression of 19,640 genes were obtained. Immunohistochemistry (IHC) data based on TCGA was downloaded from The Human Protein Atlas (https://www.proteinatlas.org/). The staining intensity was scored as negative (0), weak (1), moderate (2), or strong (3). The staining quantity was scored as 1 (≤10%), 2 (11%-50%), 3 (51%-75%), or 4 (>75%). The total immunostaining score was calculated by multiplying the staining intensity score with the quantity and ranged from 0 to 12 18.
Patients and tissue specimens
The present study was approved by the Ethics Committee of The First Hospital of Hebei Medical University (No. 2016004) and was in accordance with the principles of Declaration of Helsinki. A total of 60 pairs of human CRC tissues and adjacent normal tissues were collected from the First Hospital of Hebei Medical University (Shijiazhuang, Hebei, China). All patients were without any therapeutic invention and had signed informed consent prior to surgery. All cancer tissues were histologically confirmed as colorectal adenocarcinoma or mucinous carcinoma and were immediately preserved in liquid nitrogen within 5 min following resection and then placed at -80°C for long-term preservation.
DNA methylation assay
Genomic DNA (gDNA) was extracted from 10 pairs of human CRC tissues and adjacent normal tissues using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol. DNA quality assessment and quantification were performed using the NanoDrop 1000 (Thermo Scientific). Bisulphite conversion was performed according to the manufacturer's recommendations for the Illumina Infinium Assay (EZ DNA methylation kit, ZYMO, Orange, CA). The converted and amplified gDNA fragment was used for hybridization with the 15-mers specific capture probe (Table 2) according to the Illumina Infinium HD methylation protocol for genomic facilities (Novogene, Beijing, China). The nucleotide substrates (A/T and C/G) were labeled by different fluorescent dyes and only the probe with complementary binding of gDNA could be single base extended. Finally, iScan software was used to read and output the methylation level results according to the fluorescence.
Table 2.
Methylation sites of TRIM67 in CRC tissues
| No. | Site | Methylation | Pearson's correlation | ProbeSeq | ||
|---|---|---|---|---|---|---|
| FC | P | R | P | |||
| 1 | -1709 | 0.83 | 0.005 | 0.291 | 0.213 | AATAAACTAAACCTAACAAAATTAAAATCTTATAAAAAAAACCCCTAACC |
| 2 | -722 | 1.37 | 0.045 | -0.434 | 0.056 | AATCCCCRAAATAAATAACACTTAAACTAACAAACAAAACAATAAAAAAC |
| 3 | -678 | 1.20 | 0.011 | -0.500 | 0.025 | AAAACATCACACTACTCACAAAACATAACTACTAACTCTAACAAAATACA (U) AAAACGTCGCACTACTCGCAAAACGTAACTACTAACTCTAACGAAATACG (M) |
| 4 | -359 | 1.31 | 0.028 | -0.484 | 0.031 | TTAAAAAACTTAAAACTCRAAATATAAAACCCRTAACCRCTATCTATCTC |
| 5 | -84 | 1.64 | 0.009 | -0.543 | 0.013 | AACRCAAAACAAACAAACATTATAAAAACAAAACAAAATAATAAACTCCC |
| 6 | 23 | 1.46 | 0.013 | -0.433 | 0.056 | CAACACTACTCTACACTAAAAATAAATAACCAAAACTCAAAAAACTAACA (U) CAACGCTACTCTACGCTAAAAATAAATAACCGAAACTCGAAAAACTAACG (M) |
| 7 | 64 | 1.90 | 0.006 | -0.389 | 0.090 | AAAAAACRTACCCCTCRACTATAAAATAAACATACCCRTATAATACCCCC |
| 8 | 497 | 1.03 | 0.613 | -0.097 | 0.684 | ACAAACAAATACAAAAACCACTCAACCATAAAACTAAAATCAACACCACA (U) ACAAACAAATACGAAAACCGCTCAACCGTAAAACTAAAATCAACACCACG (M) |
| 9 | 670 | 1.25 | 0.438 | -0.161 | 0.497 | AAACCAAAAAAACAAATCAATCCCAACCACAAAAAAAAAACCCCAAAACA (U) AAACCAAAAAAACGAATCAATCCCGACCGCAAAAAAAAAACCCCAAAACG (M) |
| 10 | 706 | 1.26 | 0.262 | 0.022 | 0.928 | RACCCAAAATCAAAAACRCCRCAAAAACAACAACTAAAAACCAAAAAAAC |
| 11 | 925 | 1.94 | 0.008 | -0.284 | 0.225 | CCTTAAAATTACCCCTTAATTCCCTAACAACAAAAAACRAACCCTTATTC |
| 12 | 937 | 1.25 | 0.311 | -0.250 | 0.287 | TCCCTACAAAACCCTTAAAATTACCCCTTAATTCCCTAACAACAAAAAAC |
| 13 | 1057 | 1.67 | 0.032 | -0.103 | 0.664 | CCATTATTTAAATAAAAATTATATCTTAAATCAATACAACRACCTACCCC |
| 14 | 1119 | 1.50 | 0.041 | -0.638 | 0.002 | ATTTCTAAACAACTAACTTTTTAAAATCCCATTTTCACCCTTCTATAACA (U) ATTTCTAAACGACTAACTTTTTAAAATCCCGTTTTCACCCTTCTATAACG (M) |
| 15 | 1158 | 2.06 | 0.028 | -0.522 | 0.018 | RTAATTTTAAAAAACRACTATCAAACCATATTACCTATCCATTTCTAAAC |
Site, distance to transcription start site; FC, fold change of beta value (cancer vs. paired normal tissue); Pearson's correlation, Pearson's correlation between DNA methylation level and TRIM67 expression.
Cell culture and transfection
Human CRC cell lines HCT116, SW480, LoVo, SW620, SW1116, Caco2, SW1463, and HT29 cell lines were obtained from Prof. Jun Yu (The Chinese University of Hong Kong, Hong Kong, China). HCT116 and HT29 were cultured in McCoy's 5A medium (Gibco, Grand Island, NY, USA) and the other cells lines were cultured in DMEM medium (Gibco), supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA, USA) in a 37°C humidified incubator with 95% air and 5% CO2.
PLV-TRIM67-shRNA and negative control plasmids were obtained from Prof. Jun Yu. M98-TRIM67, M98-MAPK11, siMAPK11, and negative control plasmids were purchased from GeneCopoeia (Guangzhou, China). The plasmids were transfected into CRC cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR analysis)
Total RNA was extracted from frozen tissues and cells using TRIzol® reagent (Invitrogen) according to the manufacturer's protocol. cDNA was produced from the RNA by performing reverse transcription using a PrimeScript™ RT Reagent Kit (Perfect Real Time; Takara, Otsu, Japan). The RT-qPCR experiment was performed using AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China) according to the manufacturer's protocol. The final reaction was performed with a 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) using the following protocol: hot-start DNA polymerase activation to 95°C for 5 min; 40 cycles of 95°C for 10 sec, and 60°C for 30 sec. The specific primers were as follows: TRIM67, forward, 5'-TCCCAACTGTTTGCCACAGG-3' and reverse, 5'-AGGTTAGAACGGAACGCCTC-3'; MAPK11, forward, 5'-CGACGAGCACGTTCAATTCC-3' and reverse, 5'-TCACAGTCCTCGTTCACAGC-3'; GAPDH, forward, 5'-AGCCTCAAGATCATCAGCAATG-3' and reverse, 5'-TGTGGTCATGAGTCCTTCCACG-3'. The expression of TRIM67 and MAPK11 was normalized to GAPDH and relative expression levels were calculated using the 2-∆∆Ct method.
Western blot
Forty-eight hours after transfection, total cellular proteins were extracted using sodium dodecyl sulfate sample buffer and lysates were processed for western blot analysis. Equal amounts of total protein from each group were separated via SDS-PAGE and transferred to polyvinylidene difluoride membranes (PVDF, Millipore, Darmstadt, Germany), which were blocked with 5% skimmed milk in PBS-T for 2 h. The membranes were incubated with primary antibodies specific to TRIM67 (1:200; Abcam, Cambrige, MA, USA), MAPK11 (1:1000; Abcam), and β-actin (1:3500; Proteintech, Wuhan, China) at 4°C overnight. The membranes were then incubated with secondary DyLight fluorescent dye-conjugated antibodies (1:2500; Invitrogen) for 1 h at room temperature. Protein signals were detected and scanned by the Odyssey CLx Imaging System (LI-COR Biosciences, Lincoln, USA).
Cell proliferation and colony formation assay
For the proliferation assay, transfected cells were seeded into 96-well plates and cultured for 24 h. Then 10 μl cell counting kit-8 (CCK-8) reagent (Dojindo, Kumamoto, Japan) was added to each well according to the manufacturer's protocol 24 h. Quadruplicate OD450 values were determined by Promega GloMax Luminescence detector after 2 h incubation. For the colony formation assay, transfected cells were seeded into 10-cm plates. The visible colonies were fixed with 4% formaldehyde and stained with crystal violet after 12 days. Colonies containing more than 50 cells were counted and each condition was performed in triplicate.
Migration and invasion assays
Cell migration and invasion assays were performed using 8 µm pores (BD Biosciences, Franklin Lakes, NJ, USA). Briefly, 2×105 cells were resuspended with serum-free medium in the upper chambers of transwell plates coated with or without Matrigel (354480; BD), and medium containing 10% FBS was added to the bottom chamber. After 24 h, cells that had successfully translocated across the membrane were fixed and stained with Diff-Quick stain kit, and counted in three randomly selected fields under a light microscope.
RNA sequencing
Total RNA from transfected cells was quantified by a NanoDrop 1000 (Thermo Scientific, Wilmington, DE, USA). The quality and integrity of the total RNA were assessed by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The Illumina Xten platform was used for high-throughput next-generation sequencing by Sangon Biotech Co., Ltd (Shanghai, China). Differentially expressed genes (DEGs) were identified as such if the log2 fold change >1 or < -1 and P<0.05. Gene Ontology (GO) enrichment and enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed.
Statistical analysis
The results of normal distribution data are expressed as mean ± SD and the results of non-normal distribution data are expressed as median and quartile spacing. Student's t-test, one-way ANOVA, two-way ANOVA, and Pearson correlation analysis were used in this study. All statistical analyses were performed using GraphPad Prism 8 (GraphPad Software, USA), SPSS Statistics 21 (IBM, NY, USA). P<0.05 was considered as statistically significant.
Results
TRIM67 expression was downregulated and was associated with clinical stage, invasive depth, tumor size, lymph node metastasis, and Dukes' stage in CRC patients
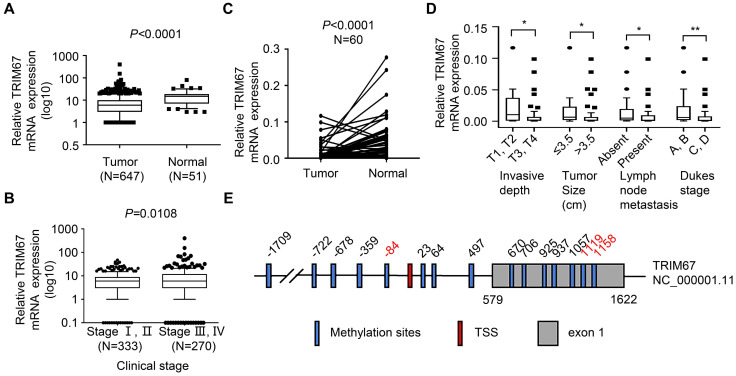
To examine TRIM67 expression levels in CRC, we first analyzed TRIM67 mRNA expression in 647 CRC tissues and 51 normal tissues from TCGA datasets. TRIM67 expression was significantly downregulated in CRC tissues compared with normal tissues. (P<0.0001; Fig. 1A). Next, we investigated whether TRIM67 expression was associated with the clinical stage of CRC patients. As shown in Figure 1B, TRIM67 expression levels of clinical stage I-II (n=333) was significantly higher than stage III-IV (n=270) CRC patients (P=0.0108) in TCGA cohort. Then we evaluated 60 pairs of CRC tissues and matched normal tissues in our cohort using RT-qPCR analysis. The results were consistent with TCGA cohort findings in that TRIM67 mRNA levels were significantly downregulated in CRC tissues than in normal tissues (P<0.0001; Fig. 1C). Further, the clinicopathological characteristics analysis of our cohort showed that TRIM67 expression was significantly correlated with invasive depth (P=0.0456), tumor size (P=0.0432), lymph node metastasis (P=0.0330), and Dukes' stage (P=0.0074; Fig. 1D), but not with sex, age, tumor location, distant metastasis, pathology, and differentiation grade (P>0.05; Table 1). In summary, higher TRIM67 expression was more likely to be detected in normal tissues, as well as earlier clinical stage, less invasive depth (T1, T2), smaller tumor size (≤3.5 cm), absent lymph node metastasis, and Dukes' stage A-B CRC tissues. These results indicated that TRIM67 may have a tumor suppressor role and act as a potential prognostic marker in CRC.
Figure 1.
TRIM67 expression is downregulated and is significantly associated with clinical stage, invasive depth, tumor size, lymph node metastasis, and Dukes' stage in human CRC tissues. A TRIM67 expression in CRC (n=647) and normal tissues (n=51) in TCGA cohort. B Relationship between TRIM67 expression and clinical stage in CRC based on TCGA cohort. C TRIM67 expression in paired CRC and normal tissues (n=60) from our cohort. D Relationship between TRIM67 expression and invasive depth, tumor size, lymph node metastasis, and Dukes' stage in CRC based on our cohort. E Schematic presentation of TRIM67 DNA methylation sites; significantly different methylation sites (FC>1.5, P<0.05) are shown in red. **P<0.01, *P<0.05.
Table 1.
Relationship between TRIM67 expression level and clinicopathological features in 60 CRC patients
| Variables | Case (n=60) | TRIM67 expression | U | P |
|---|---|---|---|---|
| Sex | 289 | 0.1118 | ||
| Male | 41 | 0.0016 (0.0005, 0.0066) | ||
| Female | 19 | 0.0061 (0.0011, 0.0235) | ||
| Age | 353 | 0.2368 | ||
| ≤65 | 36 | 0.0028 (0.0006, 0.0150) | ||
| >65 | 24 | 0.0014 (0.0005, 0.0086) | ||
| Tumor location | 430 | 0.7718 | ||
| Colon | 30 | 0.0025 (0.0005, 0.0109) | ||
| Rectum | 30 | 0.0022 (0.0004, 0.0132) | ||
| Invasive depth | 165 | 0.0456 | ||
| T1, T2 | 11 | 0.0103 (0.0008, 0.0370) | ||
| T3, T4 | 49 | 0.0017 (0.0005, 0.0069) | ||
| Tumor size (cm) | 242.5 | 0.0432 | ||
| ≤3.5 | 17 | 0.0063 (0.0017, 0.0231) | ||
| >3.5 | 43 | 0.0015 (0.0005, 0.0068) | ||
| Lymph node metastasis | 304.5 | 0.0330 | ||
| Absent | 28 | 0.0049 (0.0012, 0.0194) | ||
| Present | 32 | 0.0014 (0.0003, 0.0095) | ||
| Distant metastasis | 253.5 | 0.7653 | ||
| Absent | 49 | 0.0023 (0.0005, 0.0098) | ||
| Present | 11 | 0.0026 (0.0004, 0.0155) | ||
| Dukes | 264 | 0.0074 | ||
| A, B | 26 | 0.0059 (0.0017, 0.0238) | ||
| C, D | 34 | 0.0013 (0.0003, 0.0077) | ||
| Pathology | 158 | 0.0686 | ||
| Adenocarcinoma | 50 | 0.0017 (0.0004, 0.0095) | ||
| Mucinous carcinoma | 10 | 0.0064 (0.0015, 0.0268) | ||
| Differentiation grade | 328.5 | 0.0827 | ||
| Well | 33 | 0.0034 (0.0010, 0.0145) | ||
| Moderate, worse | 27 | 0.0014 (0.0004, 0.0068) | ||
To explore the mechanism of TRIM67 downregulation in CRC tissues, DNA methylation of the TRIM67 gene was analyzed. Fifteen DNA methylation sites are included around the transcription start site (TSS) (±2000 bp) of the TRIM67 gene (Fig. 1E). DNA hypermethylation was identified in three DNA methylation sites, and was significantly negative correlated with TRIM67 expression (FC>1.5, P<0.05; Table 2).
TRIM67 suppressed the proliferation, colony formation ability, migration, and invasion in CRC cells
To explore the role of TRIM67 in the biological behavior of CRC, we examined eight human CRC cell lines for TRIM67 expression. The results showed that TRIM67 was expressed at low levels in HCT116 cells and was highly expressed in SW480 cells (P<0.0001; Fig. 2A). Thus, TRIM67 was overexpressed in HCT116 cells using M98-TRIM67 plasmid whereas it was knocked down in SW480 using PLV-TRIM67-shRNA plasmid. Cells of the control group were transduced with negative control plasmid. Altered TRIM67 expression was examined by qRT-PCR and western blot (P<0.001; Fig. 2B and 2C).
Figure 2.
TRIM67 suppressed cell proliferation, colony formation, migration, and invasion in CRC cells. A TRIM67 expression in eight CRC cell lines was detected by RT-qPCR and western blot. B TRIM67 overexpression in HCT116 cells was detected by RT-qPCR and western blot. C TRIM67 knockdown in SW480 cells expression was detected by RT-qPCR and western blot. D Effects of TRIM67 overexpression on cell viability in HCT116 cells detected by CCK-8 assay. E Effects of TRIM67 overexpression on HCT116 colony formation assays. F Effects of TRIM67 overexpression on HCT116 cell migration assays. G Effects of TRIM67 overexpression on HCT116 cell invasion assays. H Effects of TRIM67 silencing on SW480 cell viability detected by CCK-8 assay. I Effects of TRIM67 silencing on SW480 cell colony formation assays. J Effects of TRIM67 silencing on SW480 cell migration. K Effects of TRIM67 silencing on SW480 cell invasion assays. Results show mean ± SD from three independent experiments. ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05.
Cell viability, colony formation ability, migration, and invasion was analyzed by CCK-8 assay, colony formation assay, transwell migration, and Matrigel invasion assays, respectively. These revealed that TRIM67 overexpression significantly decreased the viability (P<0.0001; Fig. 2D), colony formation (P<0.01; Fig. 2E), migration (P<0.0001; Fig. 2F), and invasion abilities (P<0.0001; Fig. 2G) of HCT116 cells compared with control cells. Conversely, TRIM67 knockdown significantly increased the viability (P<0.0001; Fig. 2H), colony formation (P<0.01; Fig. 2I), migration (P<0.001; Fig. 2J), and invasion abilities (P<0.0001; Fig. 2K) of SW480 cells compared with control cells. Therefore, we concluded that TRIM67 inhibits cell viability, colony formation ability, migration, and invasion, and has a tumor suppressor role in CRC cells.
MAPK11 is a potential downstream negative regulatory gene of TRIM67 in CRC
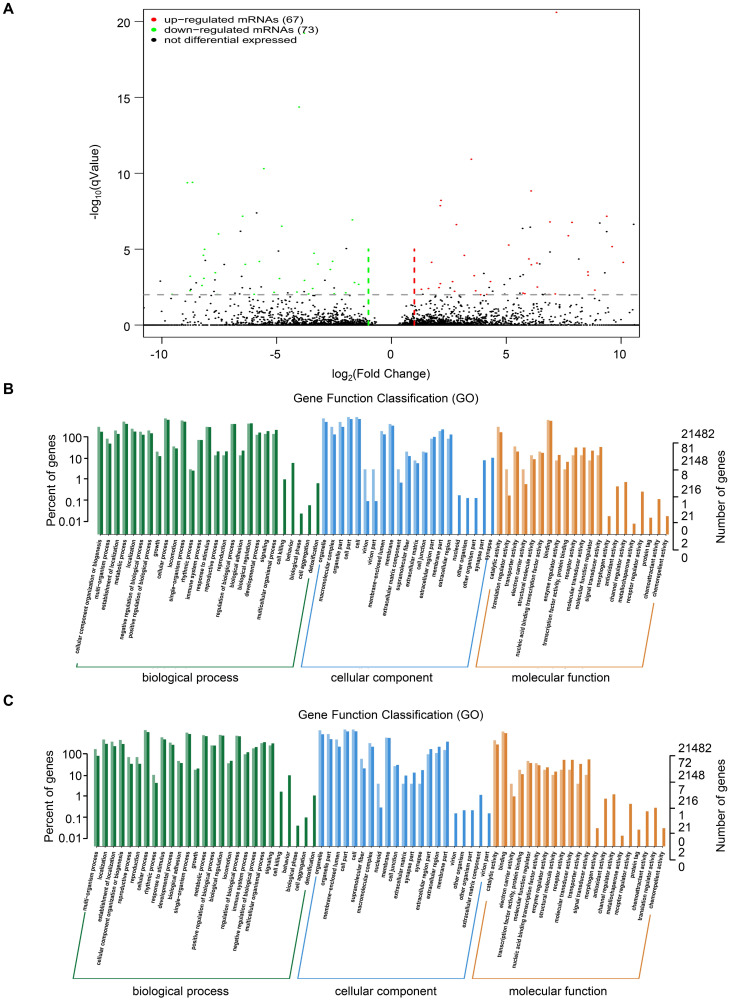
To investigate the mechanism of TRIM67 as a tumor suppressor in CRC cells, we performed RNA sequencing in HCT116 cells overexpressing TRIM67 or control vector. A total of 140 DEGs were identified, of which 67 were upregulated and 73 were downregulated (P<0.05; Fig. 3A). Detailed information of the top 20 upregulated mRNAs (log2 fold change>1, P<0.05) and top 20 downregulated mRNAs (log2 fold change< -1, P<0.05) are shown in Table 3 and 4.
Figure 3.
GO analysis of DEGs from RNA sequencing data of TRIM67 in CRC. A Volcano plot of upregulated (red spots, n=67) and downregulated mRNAs (green spots, n=73) from RNA sequencing data of TRIM67-overexpressing and control HCT116 cells. B GO analysis based on DEGs upregulated by TRIM67 from RNA sequencing data of TRIM67-overexpressing and control HCT116 cells. C GO analysis based on DEGs downregulated by TRIM67 from RNA sequencing data of TRIM67-overexpressing and control HCT116 cells.
Table 3.
Top 20 upregulated mRNAs identified by RNA sequencing (log2 fold change >1, P<0.05)
| Gene Name | Gene Description | log2 Fold Change | P |
|---|---|---|---|
| EIF3C | eukaryotic translation initiation factor 3 subunit C | 18.6087987 | 3.24E-20 |
| SDAD1 | SDA1 domain containing 1 | 17.8933647 | 4.94E-12 |
| LDLR | low density lipoprotein receptor | 17.8567578 | 1.04E-16 |
| PMS2 | PMS1 homolog 2, mismatch repair system component | 17.5701522 | 9.69E-17 |
| DDX24 | DEAD-box helicase 24 | 17.4336615 | 3.53E-12 |
| IGF2BP2 | insulin like growth factor 2 mRNA binding protein 2 | 17.3794122 | 9.43E-08 |
| MRPL48 | mitochondrial ribosomal protein L48 | 17.3358904 | 6.70E-07 |
| ARL4C | ADP ribosylation factor like GTPase 4C | 16.8662486 | 5.24E-16 |
| SMG7 | SMG7, nonsense mediated mRNA decay factor | 16.7889392 | 7.55E-15 |
| AL133352.1 | - | 16.7885144 | 4.32E-12 |
| IFI6 | interferon alpha inducible protein 6 | 16.6868835 | 1.02E-09 |
| SLC35A2 | solute carrier family 35 member A2 | 16.5691634 | 9.07E-07 |
| AC010531.1 | - | 16.3074143 | 2.91E-09 |
| ALG8 | ALG8, alpha-1,3-glucosyltransferase | 16.3050405 | 1.52E-07 |
| PRKACB | protein kinase cAMP-activated catalytic subunit beta | 16.2828954 | 1.13E-14 |
| DHPS | deoxyhypusine synthase | 16.2554958 | 2.49E-05 |
| TRAF7 | TNF receptor associated factor 7 | 16.1516508 | 4.00E-09 |
| DPP9 | dipeptidyl peptidase 9 | 16.1141761 | 2.33E-11 |
| ADGRG6 | adhesion G protein-coupled receptor G6 | 16.075702 | 2.26E-12 |
| MGA | MGA, MAX dimerization protein | 15.9977382 | 6.83E-19 |
Table 4.
Top 20 downregulated mRNAs identified by RNA sequencing (log2 fold change <-1, P<0.05)
| Gene Name | Gene Description | log2 Fold Change | P |
|---|---|---|---|
| COPB2 | coatomer protein complex subunit beta 2 | -19.363245 | 1.84E-23 |
| RTFDC1 | replication termination factor 2 domain containing 1 | -17.806038 | 6.95E-14 |
| DDX24 | DEAD-box helicase 24 | -17.59781 | 5.52E-11 |
| TPD52 | tumor protein D52 | -17.575332 | 4.96E-08 |
| ANXA5 | annexin A5 | -16.9959 | 7.47E-08 |
| RNF111 | ring finger protein 111 | -16.981478 | 2.91E-16 |
| BCKDK | branched chain ketoacid dehydrogenase kinase | -16.919981 | 2.76E-09 |
| MDH2 | malate dehydrogenase 2 | -16.81545 | 2.41E-11 |
| SRPX | sushi repeat containing protein, X-linked | -16.702299 | 8.78E-08 |
| MRPL28 | mitochondrial ribosomal protein L28 | -16.649013 | 1.42E-06 |
| FBXO9 | F-box protein 9 | -16.629698 | 4.15E-10 |
| ME2 | malic enzyme 2 | -16.584904 | 2.39E-11 |
| MAPK11 | mitogen-activated protein kinase 11 | -16.550747 | 8.84E-12 |
| GPAT2 | glycerol-3-phosphate acyltransferase 2, mitochondrial | -16.52548 | 8.52E-12 |
| BUD13 | BUD13 homolog | -16.418643 | 1.33E-06 |
| SARAF | store-operated calcium entry associated regulatory factor | -16.377437 | 6.96E-12 |
| ARFGAP2 | ADP ribosylation factor GTPase activating protein 2 | -16.308007 | 2.99E-05 |
| MOGS | mannosyl-oligosaccharide glucosidase | -16.287712 | 1.29E-05 |
| CAMTA2 | calmodulin binding transcription activator 2 | -16.262853 | 1.72E-12 |
| CCNB1IP1 | cyclin B1 interacting protein 1 | -16.248718 | 6.82E-10 |
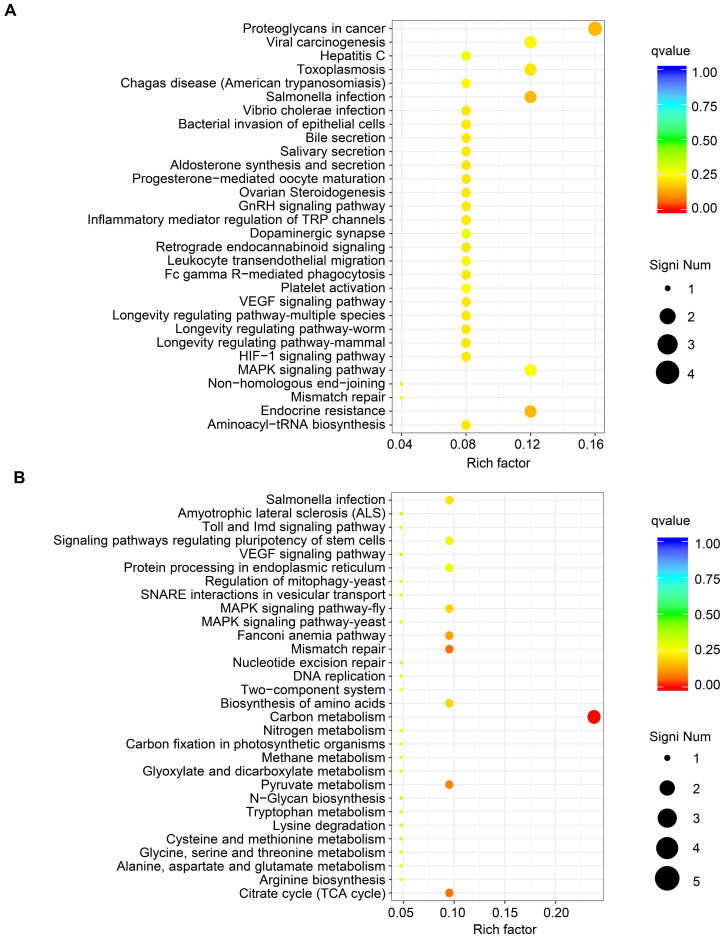
GO functional analysis of DEGs showed that the biological processes they were significantly enriched in were cellular processes, metabolic processes, developmental processes, signaling, growth, and biological adhesion. The cellular components they were most located in were cell, organelle, membrane, extracellular region, cell junction. Furthermore, the molecular functions they were involved in were binding, molecular function regulation, transporter activity, and receptor activity. (Fig. 3B and Fig. 3C). KEGG pathway analysis of upregulated mRNAs revealed that they were significantly enriched in proteoglycans in cancer and its significant symbols PRKACB, PXN, RPS6KB2, and MAPK11, VEGF signaling pathway and its significant symbols PXN and MAPK11, and Salmonella infection and its significant symbols KLC1, IFNGR2, and MAPK11 (Fig. 4A). Downregulated mRNAs were significantly enriched in carbon metabolism and its significant symbols MDH2, OGDH, PSPH, ME2, and CPS1, mismatch repair and its significant symbols RFC3 and PMS2, and MAPK signaling pathway and its significant symbols TBL1X and MAPK11 (Fig. 4B). Above all, MAPK11 (log2 fold change=-16.55, P<0.05; Table 4) was revealed to be one of the most significant downstream target genes regulated by TRIM67.
Fig 4.
KEGG pathway analysis of DEGs from RNA sequencing data of TRIM67 in CRC. A KEGG pathway analysis based on DEGs upregulated by TRIM67 from RNA sequencing data of TRIM67-overexpressing and control HCT116 cells. B KEGG pathway analysis based on DEGs downregulated by TRIM67 from RNA sequencing data of TRIM67-overexpressing and control HCT116 cells.
To investigate MAPK11 was a potential downstream negative regulatory gene of TRIM67, we performed the experiment of TRIM67 overexpression using M98-TRIM67 plasmid in HCT116 cells, and performed TRIM67 knockdown using PLV-TRIM67-shRNA plasmid in SW480 cells. RT-qPCR and western blot were used to analyze the regulation of MAPK11 by altering TRIM67 expression. As shown in Figure 5A, MAPK11 expression was downregulated in TRIM67-overexpressed HCT116 cells (P<0.05) while upregulated in TRIM67-silenced SW480 cells (P<0.01; Fig. 5B). Moreover, in our cohort, the expression of MAPK11 in cancer tissues was higher than in paired normal tissues (P=0.0017; Fig. 5C). Similarly, in TCGA cohort, MAPK11 expression was higher than in paired normal tissues (P<0.0001); representative images of MAPK11 staining are shown in Figure 5D.
Figure 5.
TRIM67 negatively regulated MAPK11 expression in CRC. A MAPK11 expression was detected by RT-qPCR and western blot in TRIM67-overexpressing HCT116 cells. B MAPK11 expression was detected by RT-qPCR and western blot in TRIM67-silenced SW480 cells. C MAPK11 expression in paired CRC and normal tissues (n=60) from our cohort was detected by RT-qPCR. D MAPK11 expression in CRC and normal tissues from TCGA cohort was detected by IHC. Results show mean ± SD from three independent experiments. ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05.
Reversing MAPK11 expression can rescue the effects of TRIM67 on CRC cell proliferation, colony formation ability, migration, and invasion
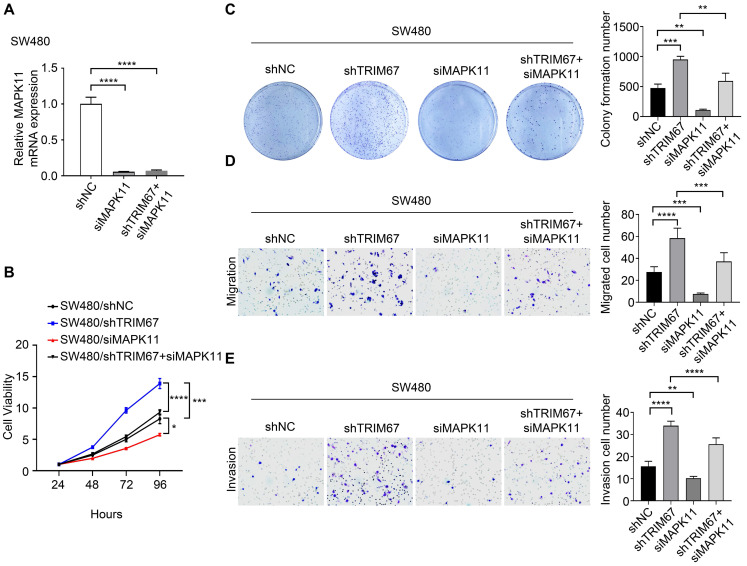
Co-overexpression of both TRIM67 and MAPK11 in HCT116 cells and co-depletion of both TRIM67 and MAPK11 in SW480 cells was performed to investigate whether MAPK11 mediates the influence of TRIM67 on the biological behavior of CRC cells. Altered MAPK11 expression was examined by qRT-PCR (P<0.0001; Fig. 6A). The results showed that overexpression of TRIM67 significantly decreased the proliferation (P<0.01; Fig. 6B), colony formation ability (P<0.0001; Fig. 6C), migration (P<0.0001; Fig. 6D), and invasion (P<0.001, Fig. 6E) of HCT116 cells, while overexpression of MAPK11 significantly increased the proliferation, colony formation ability, migration, and invasion of HCT116 cells (P<0.05; Fig. 6A-6E). Compared with overexpression of TRIM67 alone, co-overexpression of both TRIM67 and MAPK11 restored cell proliferation, colony formation ability, migration, and invasion to levels near to control cells (P<0.05; Fig. 6A-E). The depletion of TRIM67 and/or MAPK11 resulted in the opposite effects in SW480 cells (P<0.05; Fig. 7A-E). These results revealed that reversing MAPK11 expression could rescue the inhibitory effects of TRIM67 on cell proliferation, colony formation ability, migration, and invasion in CRC.
Figure 6.
MAPK11 reversed the influence of TRIM67 on cell proliferation, colony formation, migration, and invasion in HCT116 cells. A Co-regulation of TRIM67 and/or MAPK11 was detected by RT-qPCR. B Effects of co-regulation of TRIM67 and/or MAPK11 on HCT116 cell viability were detected by CCK-8 assay. C Effects of co-regulation of TRIM67 and/or MAPK11 on HCT116 cell colony formation assays. D Effects of co-regulation of TRIM67 and/or MAPK11 on HCT116 cell migration assays. E Effects of co-regulation of TRIM67 and/or MAPK11 on HCT116 cell invasion assays. Results show mean ± SD from three independent experiments. ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05.
Figure 7.
MAPK11 reversed the effect of TRIM67 on cell proliferation, colony formation, migration, and invasion in SW480 cells. TRIM67 and MAPK11 were knocked down in SW480 cells. A Co-depletion of TRIM67 and/or MAPK11 was detected by RT-qPCR. B Effects on SW480 cell viability were detected by CCK-8 assay. C Effects on SW480 colony formation were detected by colony formation assay. D Effects on SW480 cell migration were detected by cell migration assay. E Effects on SW480 cell invasion were detected by invasion assay. Results show mean ± SD from three independent experiments. ****P<0.0001, ***P<0.001, **P<0.01, *P<0.0.
Discussion
In this study, further examination of TRIM67 expression demonstrated that it was downregulated in CRC tissues compared with matched normal tissues in our cohort. Furthermore, higher TRIM67 expression was associated with earlier clinical stage and Dukes' stage, less invasive depth and lymph node metastasis, smaller tumor size. To our knowledge, this is the first study to reveal TRIM67 expression and clinicopathological features in CRC, indicating that TRIM67 may have a tumor suppressor role as well as acting as a prognostic biomarker for CRC.
Previous studies and the present study demonstrated that TRIM67 suppresses colorectal carcinogenesis and tumor growth in vivo, and plays tumor suppressor role in multiple biological functions including proliferation, colony formation ability, apoptosis, cell cycle, migration, and invasion in vitro. Mechanically, the previous study found that TRIM67 activates the p53 signaling pathway to suppress CRC initiation and progression. To gain insights into mechanism of TRIM67 as a CRC suppressor, RNA sequencing in present study revealed that TRIM67 targeted MAPK11 to play a tumor suppressor role in CRC cells. Studies have showed that MAPK signaling pathways are involved in tumor progression, cell growth, cell proliferation, and metastasis in CRC 19-22. The MAPK pathways include extracellular-regulated kinases 1 and 2 (ERK1/ERK2), c-Jun N-terminal kinases (JNKs), p38 (MAPK11-14), and extracellular-regulated kinase 5 (ERK5). p38 comprises p38β (MAPK11), p38γ (MAPK12), p38δ (MAPK13), and p38α (MAPK14), which are involved in numerous cellular functions including proliferation, apoptosis, cell cycle, autophagy, and metastases 23, 24. MAPK11 was shown to be upregulated in hepatocellular carcinoma 25 and is a target gene of miR-516a-5p sponged by circ-0001955 to facilitate hepatocellular carcinoma tumorigenesis 26. MAPK11 is also highly expressed in breast cancer cells and enhances osteoclastogenesis and bone resorption 27. In lung cancer, MAPK11 is involved in the p38 MAPK signaling pathway, and a p38-specific inhibitor decreases cell viability and enhances cisplatin sensitivity 28. These studies suggest that MAPK11 is an oncogene that functions in diverse cancers. In the present study, we found that MAPK11 was upregulated in CRC tissues compared with normal tissues, and that overexpression or depletion of TRIM67 had a negative regulatory effect on the expression of MAPK11 in CRC cell lines. Moreover, co-regulation of TRIM67 and MAPK11 expression reversed the impact of regulating TRIM67 expression alone on the biological behavior of CRC cells, indicating that MAPK11 could be a downstream negative regulatory gene of TRIM67 and might be involved in TRIM67-mediated biological behavior in CRC cells. Studies have revealed that TRIM45 inhibits the transcriptional activities of ElK-1 and AP-1 and acts as a negative transcriptional regulator in MAPK-mediated signaling 29 or functions as a member of the negative feedback loop of MAPK signaling 30. We speculated that TRIM67 might be a negative transcriptional regulator in MAPK signaling and inhibits the phosphorylation of target transcription factors, negatively regulating the expression of MAPK11 in CRC cells.
Furthermore, KEGG pathway analysis of the identified DEGs revealed that they are also enriched in other CRC-associated pathways. Proteoglycans in cancer and VEGF signaling were associated with miR-181 target genes enriched in CRC with poor prognosis 31. Sophocarpine inhibited CRC cell migration via downregulation of the MEK/ERK/VEGF pathway 32, and pyrosoda curcumin exerted inhibition of angiogenesis through modulation of VEGF signaling regulatory miRNAs 33. Defective DNA mismatch repair was associated with genomic instability, which is one of the main mechanisms in CRC 34. It was reported that the downstream genes of the above CRC-related signaling pathways have important roles in CRC. PXN was revealed to be regulated by miR-137 to promote tumor progression and metastasis in CRC 35. Another downstream gene, PSPH, was overexpressed in tumor tissues and was positively correlated with depth of invasion and distant metastasis in CRC 36. Moreover, RFC3 mutation and loss occurred in large fractions of CRC, which contributed to cancer pathogenesis by deregulating DNA repair and replication 37. According to the evidence above, we speculate that the potential downstream regulatory genes of TRIM67 in CRC might include PXN, PSPH, and RFC3, in addition to MAPK11, and might be involved in proteoglycans in cancer, VEGF, mismatch repair, or other signaling pathways to inhibit the proliferation and metastasis of CRC.
In conclusion, our data provide important insights into the tumor suppressive role of TRIM67 in CRC. We demonstrated that TRIM67 expression is correlated with clinical stage, invasive depth, tumor size, lymph node metastasis, and Dukes' stage in CRC. Moreover, TRIM67 inhibits tumor proliferation, colony formation ability, migration, and invasion via regulation of MAPK11. In future, further studies are necessary to identify direct binding sites, transcription factors, or other molecular compounds that are involved in the interaction of TRIM67 and MAPK11, as well as other potential downstream target genes and additional underlying mechanisms of TRIM67-mediated tumor suppressor functions. Besides, more analysis and clinical research are in need to clarify the functions of TRIM67 as a potential biomarker and promising therapeutic target for CRC.
Acknowledgments
This work was supported by the National Natural Science Foundation, China (grant no. 81572758); Hebei Province Key Research and Development Project (grant no. 18277741D); The Foundation for Distinguished Young Talents in Higher Education of Hebei (grant no. BJ2018042); the Graduate Innovation Funding Project Foundation, Hebei, P.R. China (grant no. CXZZBS2018079) and other Hebei Province Projects (grant no. 2019YX006A, YZ201802, LS201905, LNB201911, zh2018002, 1387, SGH201501, XH201701). The results published here are in part based upon data generated by The Cancer Genome Atlas. Information about TCGA can be found at (http://cancergenome.nih.gov), and information about The Human Protein Atlas can be found at (https://www.proteinatlas.org/). We thank Prof. Jun Yu for generously providing all the Human CRC cell lines in the present study.
Abbreviations
- CRC
colorectal cancer
- TRIM67
tripartite motif-containing 67
- TCGA
the cancer genome atlas
- MAPK
mitogen-activated protein kinase
- shRNA
short hairpin RNA
- CCK-8
cell counting kit-8
References
- 1.Bray F, Ferlay J, Soerjomataram I. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Weitz J, Koch M, Debus J. et al. Colorectal cancer. Lancet. 2004;365:153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- 3.Kohonen-Corish MRJ, Jason T, Charles C. et al. KRAS mutations and CDKN2A promoter methylation show an interactive adverse effect on survival and predict recurrence of rectal cancer. International Journal of Cancer Journal International Du Cancer. 2014;134:2820–2828. doi: 10.1002/ijc.28619. [DOI] [PubMed] [Google Scholar]
- 4.Fletcher AJ, Towers GJ. Inhibition of retroviral replication by members of the TRIM protein family. Current Topics in Microbiology & Immunology. 2013;371:29–66. doi: 10.1007/978-3-642-37765-5_2. [DOI] [PubMed] [Google Scholar]
- 5.Hatakeyama S. TRIM proteins and cancer. Nature Reviews Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 6.Maegawa H, Nakayama EE, Kuroishi A. et al. Silencing of tripartite motif protein (TRIM) 5alpha mediated anti-HIV-1 activity by truncated mutant of TRIM5alpha. Journal of Virological Methods. 2008;151:249–256. doi: 10.1016/j.jviromet.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Mandell MA, Kimura T, Jain A. et al. TRIM proteins regulate autophagy: TRIM5 is a selective autophagy receptor mediating HIV-1 restriction. Autophagy. 2014;10:2387–2388. doi: 10.4161/15548627.2014.984278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajsbaum R, García-Sastre A, Versteeg GA. TRIMmunity: The Roles of the TRIM E3-Ubiquitin Ligase Family in Innate Antiviral Immunity. Journal of Molecular Biology. 2014;426:1265–1284. doi: 10.1016/j.jmb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee OH, Lee J, Lee KH. et al. Role of the focal adhesion protein TRIM15 in colon cancer development. BBA-Molecular Cell Research. 2015;1853:409–421. doi: 10.1016/j.bbamcr.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Feng Y, Ji D, Wang Q, Qian W, Wang S. et al. TRIM27 functions as an oncogene by activating epithelial-mesenchymal transition and p-AKT in colorectal cancer. Int J Oncol. 2018;53:620–632. doi: 10.3892/ijo.2018.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu W, Xu B, Yao Y. et al. RNA interference against TRIM29 inhibits migration and invasion of colorectal cancer cells. Oncology Reports. 2016;36:1411–1418. doi: 10.3892/or.2016.4941. [DOI] [PubMed] [Google Scholar]
- 12.TRIM67 tripartite motif containing 67 [Homo sapiens (human)] https://www.ncbi.nlm.nih.gov/gene/440730.
- 13.Boyer NP, Monkiewicz C, Menon S. et al. Mammalian TRIM67 Functions in Brain Development and Behavior. Eneuro. 2018;5:ENEURO.0186–18. doi: 10.1523/ENEURO.0186-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiroaki Y, Fumihiko O, Hidehisa T. et al. TRIM67 protein negatively regulates Ras activity through degradation of 80K-H and induces neuritogenesis. Journal of Biological Chemistry. 2012;287:12050–12059. doi: 10.1074/jbc.M111.307678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Do LD, Gupton SL, Tanji K. et al. TRIM9 and TRIM67 Are New Targets in Paraneoplastic Cerebellar Degeneration. Cerebellum. 2018;18:245–254. doi: 10.1007/s12311-018-0987-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R, Chen Y, Shou T, TRIM67 promotes NF-κB pathway and cell apoptosis in GA-13315-treated lung cancer cells. Molecular medicine reports 20:2936-2944. [DOI] [PubMed]
- 17.Wang SY, Zhang YQ, Huang JZ. et al. TRIM67 Activates p53 to Suppress Colorectal Cancer Initiation and Progression. Cancer research. 2019;79:4086–4098. doi: 10.1158/0008-5472.CAN-18-3614. [DOI] [PubMed] [Google Scholar]
- 18.Wei F, Zhang T, Deng SC. et al. PD-L1 promotes colorectal cancer stem cell expansion by activating HMGA1-dependent signaling pathways. Cancer letters. 2019;450:1–13. doi: 10.1016/j.canlet.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 19.Sun H, Ou B, Zhao S. et al. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. EBioMedicine. 2019;48:236–247. doi: 10.1016/j.ebiom.2019.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stramucci L, Pranteda A, Stravato A. et al. MKK3 sustains cell proliferation and survival through p38DELTA MAPK activation in colorectal cancer. Cell death & disease. 2019;10:1–13. doi: 10.1038/s41419-019-2083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi XP, Zhu M, Gong ZY. et al. Homoharringtonine suppresses LoVo cell growth by inhibiting EphB4 and the PI3K/AKT and MAPK/EKR1/2 signaling pathways. Food and chemical toxicology. 2020;136:110960. doi: 10.1016/j.fct.2019.110960. [DOI] [PubMed] [Google Scholar]
- 22.Koveitypour Z, Panahi F, Vakilian M. et al. Signaling pathways involved in colorectal cancer progression. Cell & bioscience. 2019;9:1–14. doi: 10.1186/s13578-019-0361-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Browne AJ, Göbel A, Thiele S, p38 MAPK regulates the Wnt inhibitor Dickkopf-1 in osteotropic prostate cancer cells. Cell Death & Disease. 7: e2119. [DOI] [PMC free article] [PubMed]
- 24.Zhong W, Zhu HC, Sheng FG. et al. Activation of the MAPK11/12/13/14 (p38 MAPK) pathway regulates the transcription of autophagy genes in response to oxidative stress induced by a novel copper complex in HeLa cells. Autophagy. 2014;10:1285–1300. doi: 10.4161/auto.28789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang Y, Li WW, Yang BW. et al. Aryl hydrocarbon receptor nuclear translocator is associated with tumor growth and progression of hepatocellular carcinoma. International journal of cancer. 2012;130:1745–1754. doi: 10.1002/ijc.26166. [DOI] [PubMed] [Google Scholar]
- 26.Yao ZC, Xu RY, Yuan L. et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell death & disease. 2019;10:1–12. doi: 10.1038/s41419-019-2176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He ZM, He J, Liu ZQ. et al. MAPK11 in breast cancer cells enhances osteoclastogenesis and bone resorption. Biochimie. 2014;106:24–32. doi: 10.1016/j.biochi.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Planchard D, Valérie Camara-Clayette, Dorvault N. et al. p38 Mitogen-activated protein kinase signaling, ERCC1 expression, and viability of lung cancer cells from never or light smoker patients. Cancer. 2012;118:5015–5025. doi: 10.1002/cncr.27510. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Li Y, Qi X, Yuan W, Ai J, Zhu C, TRIM45, a novel human RBCC/TRIM protein, inhibits transcriptional activities of ElK-1 and AP-1. Biochemical & Biophysical Research Communications. 323: 9-16. [DOI] [PubMed]
- 30.Sato T, Takahashi H, Hatakeyama S. et al. The TRIM-FLMN protein TRIM45 directly interacts with RACK1 and negatively regulates PKC-mediated signaling pathway. Oncogene. 2015;34:1280–1291. doi: 10.1038/onc.2014.68. [DOI] [PubMed] [Google Scholar]
- 31.Peng Q, Yao W, Yu C. et al. Identification of microRNA-181 as a promising biomarker for predicting the poor survival in colorectal cancer. Cancer medicine. 2019;8:5995–6009. doi: 10.1002/cam4.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Wang T, Zhu L. et al. Sophocarpine Inhibits Tumorgenesis of Colorectal Cancer via Downregulation of MEK/ERK/VEGF Pathway. Biological & pharmaceutical bulletin. 2019;42:1830–1838. doi: 10.1248/bpb.b19-00353. [DOI] [PubMed] [Google Scholar]
- 33.Moradi-Marjaneh R, Hassanian SM, Rahmani F. et al. Phytosomal Curcumin Elicits Anti-tumor Properties Through Suppression of Angiogenesis, Cell Proliferation and Induction of Oxidative Stress in Colorectal Cancer. Current pharmaceutical design. 2018;24:4626–4638. doi: 10.2174/1381612825666190110145151. [DOI] [PubMed] [Google Scholar]
- 34.Arends MJ. Pathways of Colorectal Carcinogenesis. Appl Immunohistochem Mol Morphol. 2013;21:97–102. doi: 10.1097/PAI.0b013e31827ea79e. [DOI] [PubMed] [Google Scholar]
- 35.Chen DL, Wang DS, Wu WJ. et al. Overexpression of paxillin induced by miR-137 suppression promotes tumor progression and metastasis in colorectal cancer. Carcinogenesis. 2013;34:803–811. doi: 10.1093/carcin/bgs400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato K, Masuda T, Hu Q. et al. Phosphoserine Phosphatase Is a Novel Prognostic Biomarker on Chromosome 7 in Colorectal Cancer. Anticancer research. 2017;37:2365–2371. doi: 10.21873/anticanres.11574. [DOI] [PubMed] [Google Scholar]
- 37.Kim YR, Song SY, Kim SS. et al. Mutational and expressional analysis of RFC3, a clamp loader in DNA replication, in gastric and colorectal cancers. Human pathology. 2010;41:1431–1437. doi: 10.1016/j.humpath.2010.03.006. [DOI] [PubMed] [Google Scholar]