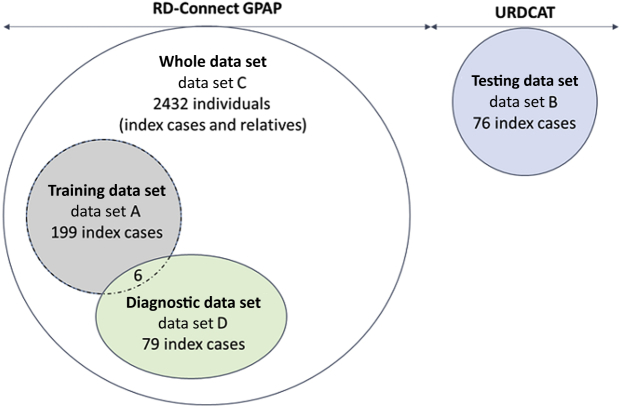
Figure 1.
Description of the different data sets used in this study. Data sets used to train, test, and evaluate the model described in this study. Data set A, referred to as training data set, includes 199 index cases for which presence or absence of consanguinity was determined by kinship analysis and was used to define the logistic regression model. Data set B, referred to as testing data set, includes 76 index cases from the Undiagnosed Rare Disease Program of Catalonia (URDCAT) for which the presence or absence of consanguinity status was determined by kinship analysis and was used to test our model. Data set C, referred to as whole data set, includes 2432 individuals (index cases and relatives) from the Rare Disease (RD)–Connect Genome-Phenome Analysis Platform (GPAP) to which our model was applied. Data set D, referred to as diagnostic data set, includes 79 index cases in which genomic data were combined with run of homozygosity results to identify the pathogenic variants responsible for different types of rare disease.