Abstract
Background—
Cytokines and growth factors have been implicated in the initiation and propagation of vascular disease. Observational studies have shown associations of their circulating levels with stroke. Our objective was to explore whether genetically determined circulating levels of cytokines and growth factors are associated with stroke and its etiologic subtypes by conducting a two-sample Mendelian randomization (MR) study.
Methods—
Genetic instruments for 41 cytokines and growth factors were obtained from a genome-wide association study (GWAS) of 8,293 healthy adults. Their associations with stroke and stroke subtypes were evaluated in the MEGASTROKE GWAS dataset (67,162 cases; 454,450 controls) applying inverse-variance-weighted meta-analysis, weighted-median analysis, MR-Egger regression, and multivariable MR. The UK Biobank cohort was used as an independent validation sample (4,985 cases; 364,434 controls). Genetic instruments for monocyte chemoattractant protein-1 (MCP-1/CCL2) were further tested for association with etiologically related vascular traits using publicly available GWAS data.
Results—
Genetic predisposition to higher MCP-1 levels was associated with higher risk of any stroke (OR per 1-SD increase: 1.06, 95% CI: 1.02–1.09, p=0.0009), any ischemic stroke (OR: 1.06, 95% CI: 1.02–1.10, p=0.002), large artery stroke (OR: 1.19, 95% CI: 1.09–1.30, p=0.0002) and cardioembolic stroke (OR: 1.14, 95% CI: 1.06–1.23, p=0.0004), but not with small vessel stroke or intracerebral hemorrhage. The results were stable in sensitivity analyses and remained significant after adjustment for cardiovascular risk factors. Analyses in the UK Biobank showed similar associations for available phenotypes (any stroke: OR: 1.08, 95% CI: 0.99–1.17, p=0.09; any ischemic stroke: OR: 1.07, 95% CI: 0.97–1.18, p=0.17). Genetically determined higher MCP-1 levels were further associated with coronary artery disease (OR: 1.04, 95% CI: 1.00–1.08, p=0.04) and myocardial infarction (OR: 1.05, 95% CI: 1.01–1.09, p=0.02), but not with atrial fibrillation. A meta-analysis of observational studies showed higher circulating MCP-1 levels in stroke patients compared to controls.
Conclusions—
Genetic predisposition to elevated circulating levels of MCP-1 is associated with higher risk of stroke, particularly with large artery stroke and cardioembolic stroke. Whether targeting MCP-1 or its receptors can lower stroke incidence requires further study.
Keywords: MCP-1, CCL2, inflammation, cytokines, atherosclerosis, stroke, Mendelian randomization, genetics, human
Twitter summary:
Mendelian randomization identifies genetic predisposition to high MCP-1 circulating levels as a risk factor for stroke
INTRODUCTION
Stroke is the leading cause of long-term disability and the second most common cause of death world-wide1, 2 with a growing burden on global health.3 Inflammatory mechanisms have been implicated in stroke and etiologic stroke subtypes,4–6 and specifically demonstrated for large artery atherosclerotic stroke.4, 5 Cytokines and growth factors regulate the inflammatory response4 and thus may serve as targets for cardiovascular disease prevention.7 Indeed, the CANTOS trial recently demonstrated the potential of targeting specific inflammatory cytokines in reducing vascular endpoints.8
Few studies have investigated associations between circulating levels of inflammatory cytokines and risk of stroke. Levels of IL-1β and IL-6 were found to be associated with incident and recurrent ischemic stroke.4 However, these associations derived from observational studies preclude conclusions about causal relationships because of possible confounding and reverse causation.9 Also, associations with etiologic stroke subtypes were not investigated in depth.4 Hence, the potential causative role of individual cytokines in determining stroke risk remains elusive. Developing meaningful strategies for stroke prevention will require defining these relationships.10
Mendelian randomization (MR) aims to overcome the limitations of conventional epidemiologic studies with respect to confounding and reverse causation. By using genetic variants as instrumental variables for a trait, MR enables an investigation of associations independent of the conventional biases accompanying observational studies.11 A recent genome-wide association study (GWAS) in 8,293 healthy subjects of Finnish ancestry identified multiple common genetic variants that influence circulating levels of 41 cytokines and growth factors (referred to hereafter as ‘cytokines’ for simplicity),12 thus providing comprehensive data on genetic determinants of circulating inflammatory biomarkers.12
Here, by leveraging data from this recent GWAS on cytokines12 and the largest GWAS meta-analysis on stroke and stroke subtypes to date,13 we implemented a two-sample MR study to: (i) explore the associations between genetic predisposition to higher or lower circulating cytokine levels with risk of any stroke; (ii) evaluate specific associations with ischemic stroke and its major etiologic subtypes (large artery stroke, cardioembolic stroke, and small vessel stroke), as well as with intracerebral hemorrhage; (iii) validate these findings in UK Biobank as an independent cohort; (iv) compare the MR associations to estimates of association derived from meta-analyses of observational studies and (v) examine the association with etiologically related cardiovascular outcomes including coronary artery disease (CAD), myocardial infarction (MI), and atrial fibrillation (AF).
METHODS
Access to publicly available data
The analyses for this study were based on publicly available summary statistics from GWAS Consortia. The web-links for downloading the data are provided in Supplemental Table 1 along with descriptive characteristics of the Consortia. The retrieved summary data for the current analysis and the code script are available upon reasonable request to the corresponding author. As all analyses have been based on publicly available summary statistics and not individual-level data, no ethical approval from an institutional review board was required.
Study design and data sources
The overall design of this study is displayed in Figure 1. Supplemental Table 1 summarizes our data sources for this MR study. The genetic instruments were taken from publicly available summary statistics.12 For each of the 41 cytokines (full list provided in Supplemental Table 2) we selected single nucleotide polymorphisms (SNPs) associated with its circulating levels at a significance threshold of a false discovery rate (FDR) <5%.14 To avoid bias by selection of false positive instruments, we performed additional analyses using a genome-wide threshold of significance (p <5×10−8). After extracting the summary statistics for significant SNPs, we pruned all SNPs in linkage disequilibrium (LD; r2 <0.1 in the European 1000G reference panel) retaining SNPs with the lowest p-value as independents instrument. We identified 698 SNPs not in LD to be significantly associated with circulating cytokine levels; 615 of them were also available in the MEGASTROKE dataset. To avoid use of pleiotropic instruments we excluded 126 SNPs that were associated with levels of more than one cytokine15 leaving 489 SNPs as the final instruments. These instruments related to the circulating levels of 23 cytokines, whereas for 18 cytokines no SNPs associated with their circulating levels at a significance level of FDR <5% could be identified.
Figure 1. Schematic representation of the study design.
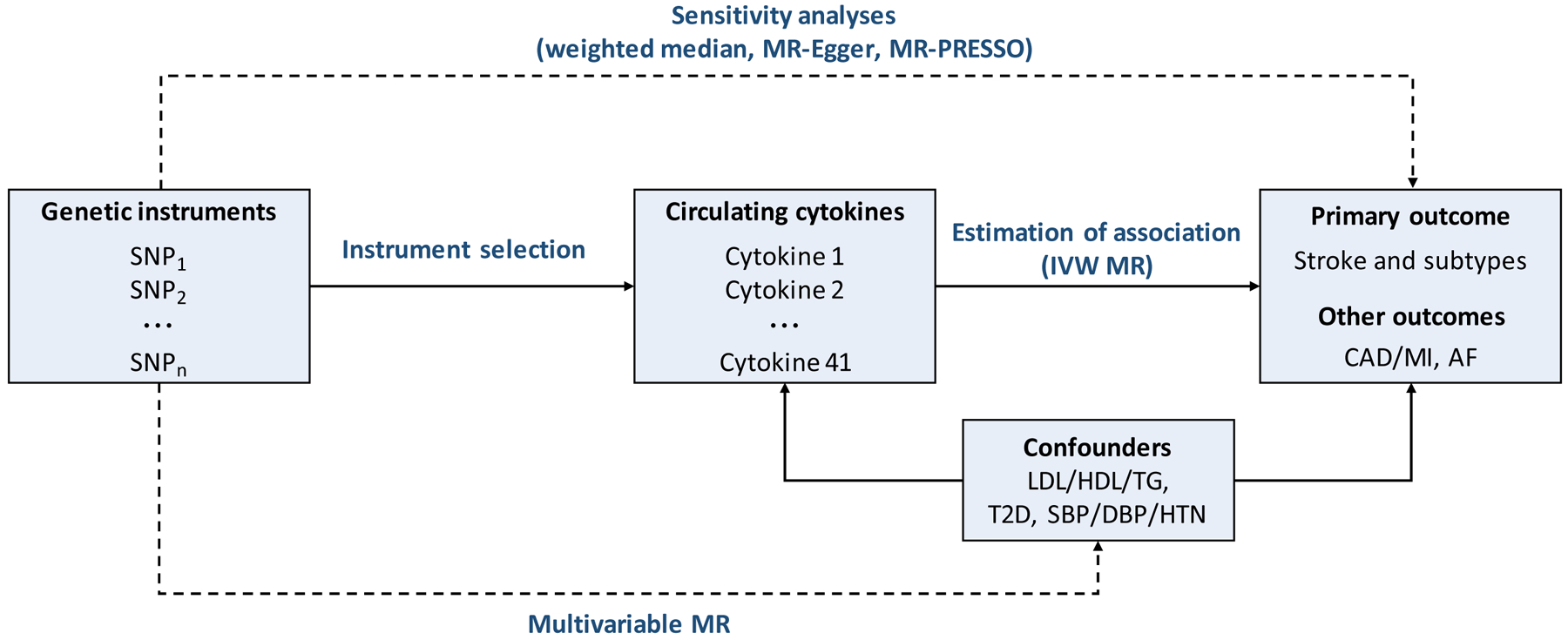
Methods used to test for associations and for violations of the Mendelian randomization assumptions (dashed lines). AF, atrial fibrillation; CAD, coronary artery disease; DBP, diastolic blood pressure; HDL, high-density lipoprotein cholesterol; HTN, hypertension; IVW, inverse-variance weighted; LDL, low-density lipoprotein cholesterol; MI, myocardial infarction; MR: Mendelian randomization; MR-PRESSO: Mendelian Randomization Pleiotropy RESidual Sum and Outlier; SBP, systolic blood pressure; SNP, Single-nucleotide polymorphism; T2D. type 2 diabetes mellitus; TG, triglycerides.
The primary outcomes for this study were any stroke, any ischemic stroke, etiologic ischemic stroke subtypes defined by TOAST criteria (large artery stroke, cardioembolic stroke, and small vessel stroke),16 and intracerebral hemorrhage. We extracted estimates for the associations of the selected instruments with any stroke, any ischemic stroke and its subtypes from the MEGASTROKE multi-ancestry GWAS dataset (67,162 cases; 454,450 controls).13 Sensitivity analyses restricted to individuals of European ancestry (40,528 cases; 445,396 controls) were conducted, to minimize ancestral mismatch with the Finnish population used for the discovery GWAS on cytokines.12 For intracerebral hemorrhage, we extracted data from publicly available summary statistics of a GWAS meta-analysis on 1,545 cases and 1,481 controls of European ancestry.17
We computed F-statistics to quantify the strength of the selected instruments18 and performed power calculations.19 The F-statistic for the 489 instrument SNPs ranged from 17 to 789 (Supplemental Table 3), well above the threshold of F >10 typically recommended for MR analyses.20 Based on the sample size of MEGASTROKE, there was >80% power to detect significant associations with any stroke and any ischemic stroke for 18 of 23 cytokines at an effect size (OR [odds ratio]) of 1.10. Power was lower for the remaining 5 cytokines and for sub-analyses for ischemic stroke subtypes and intracerebral hemorrhage (Supplemental Table 3).
For validation of significant associations in MEGASTROKE, we used the UK Biobank dataset as detailed in the Supplemental Methods. We included cases of prevalent and incident stroke. Cases with an unconfirmed self-reported diagnosis of stroke were excluded from the analysis. The final sample size consisted of 369,419 individuals, including 4,985 cases with any stroke and 3,628 cases with any ischemic stroke. No data were available on ischemic stroke subtypes.
Cytokines that were significantly associated with stroke were subsequently explored for an association with etiologically related vascular outcomes. Publicly available summary statistics were extracted from the CARDIoGRAMplusC4D Consortium for CAD and MI (60,801 CAD and 43,676 MI cases; 123,504 controls),21 and the AFGen Consortium for AF (17,931 cases; 115,142 controls).22
Statistical analysis
After extraction of data and harmonization of the effect alleles across GWASs, we computed individual MR estimates and standard errors from the SNP-cytokine and SNP-outcome associations using the Wald estimator and the Delta method that weight all estimates based on the magnitude of the SNP-cytokine association.23 The MR association between each cytokine and stroke was estimated after pooling individual SNP MR estimates using fixed-effects inverse-variance weighted (IVW) meta-analysis.23 Statistical significance for the MR associations with stroke was set at a p-value corrected for multiple comparisons (based on number of cytokines) using the Bonferroni method. We further report on results corrected for both the number of cytokines and the number of examined phenotypes. A p <0.05 but above the Bonferroni-corrected threshold was considered as suggestive for association. The IVW MR approach assumes that instruments affect the outcome only through the exposure under consideration, and not by some alternative pathway.23 Any violation of this assumption would represent horizontal pleiotropy of the instrument and could introduce bias to the MR estimate. In the absence of any such horizontal pleiotropy, there would not be any expected heterogeneity in the MR estimates obtained from different instruments. As such, heterogeneity markers (I2 >25% or Cochran Q-derived p <0.05) from the IVW MR were used as indicators of possible horizontal pleiotropy.24
For cytokines showing either significant or suggestive associations or significant heterogeneity in the primary IVW MR analysis, we conducted additional sensitivity analyses that vary in their underlying assumptions regarding the presence of pleiotropic genetic variants that may be associated with the outcome independently of the exposure. Particularly, we used MR-Egger regression, which requires that the strengths of the instruments are independent of their direct associations with the outcome,25 and the weighted median method, which requires that at least half of the information for the MR analysis comes from valid instruments.26 We used the intercept obtained from the MR-Egger regression as a measure of directional pleiotropy (p <0.05 was considered significant),25 and also tested for outlier SNPs using MR-PRESSO.27
To generate MR estimates unaffected by the presence of pleiotropic pathways acting through cardiovascular risk factors, we performed regression-based multivariable MR with summary genetic association estimates28 that adjusted for the genetic association of instruments with circulating lipid levels (LDL cholesterol, HDL cholesterol, triglycerides), type 2 diabetes (T2D), and blood pressure measurements (systolic and diastolic blood pressure, hypertension). Genetic association estimates for these phenotypes were extracted from the GLGC consortium,29 the DIAGRAM consortium,30 and the UK Biobank GWAS published by the Neale lab (https://sites.google.com/broadinstitute.org/ukbbgwasresults), respectively.
Instrument SNPs for cytokines showing significant associations with stroke were mapped to the nearest gene using the GRCh37/hg19 reference genome. We used the STRING database31 to look for protein-protein interactions between gene products and the cytokines and identified interacting subnetworks. As a sensitivity analysis and to gain further insight into the biological processes involved in the examined associations, we performed IVW MR analysis with SNPs restricted to the specific subnetworks.
The GWAS used to select cytokine instruments included no replication and its estimates of association were further adjusted for BMI, besides age and sex.12 As a sensitivity analysis for bias that may be introduced by this BMI adjustment,32 we also calculated an unweighted allele score for any cytokines demonstrating a significant association in our main IVW MR analysis.33 Such an unweighted allele score may offer evidence of a causal effect of the exposure on the outcome without suffering from bias in the genetic association estimates for the exposure, although this is at the cost of not being able to estimate the magnitude of any such effect.33 Statistical was analyses were conducted in Stata 13.1 (StataCorp).
Meta-analysis of observational studies
For the cytokines that showed significant associations with stroke in MR, we performed a meta-analysis of observational studies. We searched Medline until December 10, 2017 (search strategy is available in the Supplemental Methods), for case-control studies comparing the circulating cytokine levels between stroke patients and controls, and cohort studies exploring the association of baseline levels with incident or recurrent stroke. We extracted relevant data and applied random-effects meta-analyses for Hazard ratios (cohort studies) or standardized mean differences (case-control studies). We evaluated heterogeneity with the I2 and the Cochran Q.
RESULTS
Genetically determined circulating levels of cytokines and risk of stroke
The primary results of the MR analyses for the 23 cytokines are presented in Figure 2. Following Bonferroni correction for testing multiple cytokines (p <0.05/23=2.2×10−3), the only cytokine showing statistically significant associations with stroke was the CC chemokine monocyte chemoattractant protein-1 (MCP-1/CCL2). As depicted in Figure 3A and Supplemental Figure 1, genetically determined higher circulating MCP-1 levels (1-SD increase) were associated with 6% higher odds for both any stroke (OR: 1.06, 95%CI: 1.021.09, p=9×10−4; 523,047 individuals; 66,856 cases) and any ischemic stroke (OR: 1.06, 95%CI: 1.02–1.10, p=1.8×10−3; 511,551 individuals; 60,341 cases) in MR analyses. Corresponding analyses for ischemic stroke subtypes revealed significant associations for large artery stroke (OR: 1.19, 95%CI: 1.09–1.30, p=1.7×10−4; 245,201 individuals; 6,688 cases) and cardioembolic stroke (OR: 1.14, 95%CI: 1.06–1.23, p=3.5×10−4; 361,858 individuals; 9,006 cases), but not for small vessel stroke (OR: 1.03, 95%CI: 0.95–1.11, p=0.50; 298,777 individuals; 11,710 cases). In addition, we found no significant association of genetically determined MCP-1 levels with intracerebral hemorrhage (OR: 1.24, 95%CI: 0.94–1.64, p=0.13), although this might be related to the lower sample size (3,026 individuals; 1,545 cases). Importantly, the results for large artery stroke and cardioembolic stroke remained significant when further correcting for both the number of examined cytokines and the number of examined phenotypes (p <0.05/138=3.6×10−4; Figure 2). Sub-analyses restricted to lobar (OR: 1.25, 95%CI: 0.88–1.79, p=0.22; 2,145 individuals; 664 cases), and nonlobar intracerebral hemorrhage (OR: 1.03, 95%CI: 0.72–1.49, p=0.16; 2,362 individuals; 881 cases) also showed no significant associations with genetically determined MCP-1 levels. The individual SNPs associated with MCP-1 levels explained 14.7% of the variance of MCP-1 levels (Supplemental Table 3) and are presented in Supplemental Table 4.
Figure 2. Mendelian randomization associations of circulating cytokine and growth factor levels with stroke and stroke subtypes.
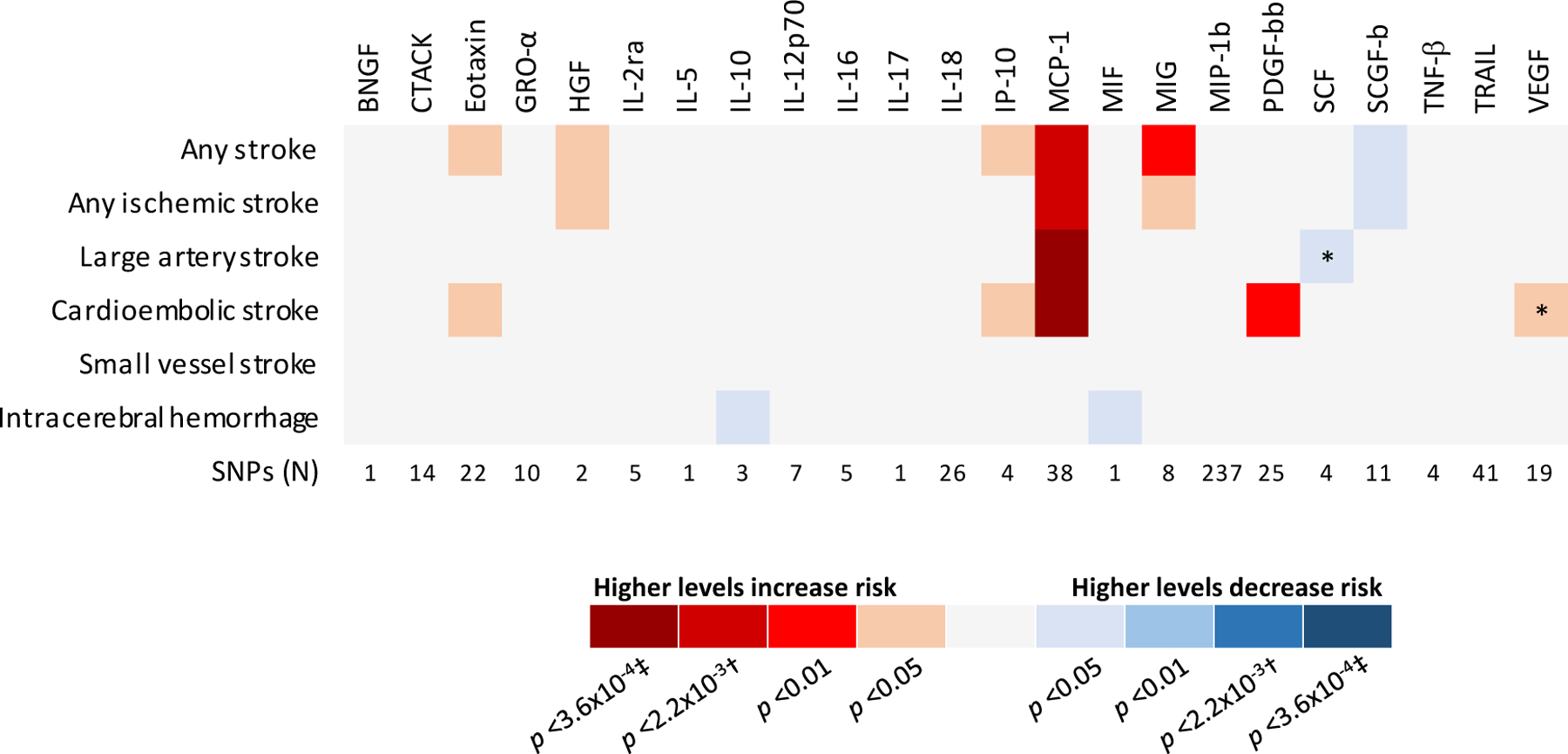
Shown are the results derived from the fixed-effects inverse-variance weighted (IVW) meta-analysis.
* Significant heterogeneity (I2>25% or Cochran Q-derived p <0.05)
† Bonferroni-corrected threshold for number of tested cytokines
‡ Bonferroni-corrected threshold for number of cytokines and number of phenotypes
Figure 3. Mendelian randomization analysis for circulating MCP-1 levels and risk of stroke.
(A) MR-derived associations between genetically determined circulating MCP-1 levels (1-SD increase) and risk of any stroke and stroke subtypes. (B) Associations between genetically determined circulating MCP-1 levels and risk of large artery (left) and cardioembolic (right) stroke based on different MR methods. I2 refers to heterogeneity in the Mendelian randomization analysis (inverse-variance weighted method). CI, confidence intervals; IVW, inverse-variance weighted; OR, Odds Ratio; SNP, single nucleotide polymorphism.
There was no evidence for heterogeneity in any of the MCP-1 associations as measured by I2 and Cochran Q (Figure 3A) and no outlier SNPs were detected with the MR-PRESSO method. Also, there was no indication for directional pleiotropy effects as assessed by the MR-Egger intercept (any stroke, p=0.41; any ischemic stroke, p=0.39; large artery stroke, p=0.98; cardioembolic stroke, p=0.67; small vessel stroke, p=0.70; intracerebral hemorrhage, p=0.94). The weighted median estimator and the MR-Egger regression analysis provided estimates of the same magnitude as the fixed-effects IVW meta-analysis for large artery stroke (OR: 1.22, 95%CI: 1.07–1.40, p=2×10−3 and OR: 1.19, 95%CI: 0.93–1.53, p=0.13, respectively) and cardioembolic stroke (OR: 1.13, 95%CI: 1.01–1.27, p=0.04 and OR: 1.21, 95%CI: 0.96–1.53, p=0.09, respectively, Figure 3B); although with wider confidence intervals as would be expected given the lower statistical power of these approaches.25, 26 Use of an unweighted allele score for the MCP-1 instrument SNPs also showed statistically significant associations with risk of large artery (p=1.5×10−4) and cardioembolic stroke (p=2.8×10−4). The significant association between MCP-1 and outcomes was retained both when restricting the analysis to individuals of European ancestry (Supplemental Figure 2), and when applying the more conservative threshold of p <5×10−8 for instrument selection (Supplemental Figure 3).
To explore whether the MR association between genetically determined MCP-1 levels and stroke was attributable through pleiotropic pathways relating to cardiovascular risk factors, we conducted multivariable MR analysis adjusting for circulating lipid levels, T2D, and blood pressure. The results remained stable regardless of the model (unadjusted, single or fully-adjusted model), thus supporting an independent association between MCP-1 levels and stroke and stroke subtypes (Table 1).
Table 1.
Multivariable Mendelian randomization associations between circulating MCP-1 levels and risk of stroke and its subtypes adjusting for cardiovascular risk factors.
| Any stroke | Any ischemic stroke | Large artery stroke | Cardioembolic stroke | Small vessel stroke | Intracerebral hemorrhage | |
|---|---|---|---|---|---|---|
| N sample | 523,047 | 511,551 | 245,201 | 361,858 | 298,777 | 3,026 |
| N cases | 66,856 | 60,341 | 6,688 | 9,006 | 11,710 | 1,545 |
| Unadjusted model | 1.06 (1.02–1.09) | 1.06 (1.02–1.10) | 1.19 (1.09–1.30) | 1.14 (1.06–1.23) | 1.03 (0.95–1.11) | 1.24 (0.94–1.64) |
| Adjusted for T2D | 1.07 (1.03–1.11) | 1.07 (1.03–1.11) | 1.22 (1.12–1.33) | 1.17 (1.08–1.27) | 1.03 (0.97–1.10) | 1.06 (0.94–1.20) |
| Adjusted for LDL-C | 1.06 (1.02–1.10) | 1.06 (1.02–1.11) | 1.20 (1.10–1.31) | 1.16 (1.06–1.24) | 1.03 (0.98–1.09) | 1.26 (0.93–1.71) |
| Adjusted for HDL-C | 1.07 (1.03–1.11) | 1.07 (1.02–1.11) | 1.21 (1.11–1.33) | 1.15 (1.06–1.25) | 1.04 (0.97–1.10) | 1.27 (0.94–1.72) |
| Adjusted for TG | 1.06 (1.02–1.10) | 1.06 (1.02–1.10) | 1.19 (1.09–1.30) | 1.16 (1.06–1.26) | 1.03 (0.97–1.10) | 1.28 (0.94–1.73) |
| Adjusted for SBP | 1.08 (1.04–1.12) | 1.09 (1.05–1.14) | 1.23 (1.12–1.35) | 1.20 (1.10–1.32) | 1.03 (0.96–1.11) | 1.81 (1.13–1.90) |
| Adjusted for DBP | 1.08 (1.04–1.13) | 1.09 (1.05–1.14) | 1.22 (1.11–1.34) | 1.20 (1.10–1.32) | 1.04 (0.96–1.11) | 1.53 (0.89–2.65) |
| Adjusted for HTN | 1.07 (1.03–1.11) | 1.07 (1.03–1.11) | 1.19 (1.09–1.29) | 1.18 (1.08–1.29) | 1.03 (0.95–1.11) | 1.03 (0.93–1.14) |
| Fully-adjusted model (T2D, LDL-C*, SBP †) | 1.08 (1.03–1.12) | 1.09 (1.04–1.13) | 1.23 (1.11–1.35) | 1.20 (1.10–1.32) | 1.04 (0.97–1.12) | 1.06 (0.92–1.21) |
The results are presented as Odds Ratios (95% Confidence Intervals) for the effect of 1 standard deviation increase in MCP-1 levels.
restricted to LDL-C to avoid collinearity with HDL-C and TG levels.
restricted to SBP to avoid collinearity with DBP and HTN.
DBP, diastolic blood pressure; HDL-C, high-density lipoprotein cholesterol; HTN, hypertension; LDL-C, low-density lipoprotein cholesterol; SBP: systolic blood pressure; T2D, type 2 diabetes mellitus; TG, triglycerides.
None of the genetic instruments for MCP-1 was within or close to the MCP1 gene. Assessing genes closest to the instruments for MCP-1 we noted that several of them encoded proteins that show a biological relationship with MCP-1, e.g. CCR2 the main receptor for MCP1 (Supplemental Table 4). To minimize the risk of using nonspecific instruments that might exert pleiotropic effects we performed an additional sensitivity analysis focusing on instruments in the vicinity of these genes. Using the STRING database, we found the chemokine receptors CCR2, CCR1, CCR3, and CCR9, the chemokine binding protein CCBP2, and the receptor of the complement C5a (C5aR1) to integrate into a subnetwork of established interactions with MCP-1 (Supplemental Figure 4A). Restricting the MR analysis to the respective SNPs, resulted in significant estimates of association for large artery (OR per 1-SD increase in MCP-1 levels: 1.25, 95%CI: 1.08–1.45, p=2×10−3) and cardioembolic stroke (OR: 1.21, 95%CI: 1.07–1.37, p=3×10−3), as well as intracerebral hemorrhage (OR: 2.19, 95%CI: 1.30–3.69, p=3×10−3) (Supplemental Figure 4B).
Several other cytokines not reaching the Bonferroni-corrected threshold showed suggestive (p <0.05) associations with risk of stroke in MR analyses: genetic predisposition to higher levels of eotaxin, IP-10, MIG, PDGF-bb, and VEGF were associated with an higher risk of stroke whereas predisposition to higher levels of SCF and SCGF-b were associated with lower risk of stroke (Figure 2).
Genetically determined circulating levels of MCP-1 and risk of stroke in UK Biobank
We next explored the MR association between genetically determined MCP-1 levels and risk of any stroke and risk of any ischemic stroke in the independent UK Biobank sample and meta-analyzed the MEGASTROKE and UK Biobank data (Figure 4A and Supplemental Figure 5). Estimates of association in UK Biobank were similar to MEGASTROKE for any stroke (OR per 1-SD increase: 1.08, 95%CI: 0.99–1.17, p=0.09; 369,419 individuals, 4,985 cases) and any ischemic stroke (OR: 1.07, 95%CI: 0.97–1.18, p=0.17; 369,419, 3,628 cases), but did not reach statistical significance. Genetically elevated circulating MCP-1 levels were significantly associated with both any stroke (OR: 1.06, 95%CI: 1.03–1.09, p=2×10−4) and any ischemic stroke (OR: 1.06, 95%CI: 1.03–1.10, p=7×10−4) in the meta-analysis of MEGASTROKE and UK Biobank
Figure 4. Associations between circulating MCP-1 levels and risk of stroke in Mendelian randomization and in observational studies.
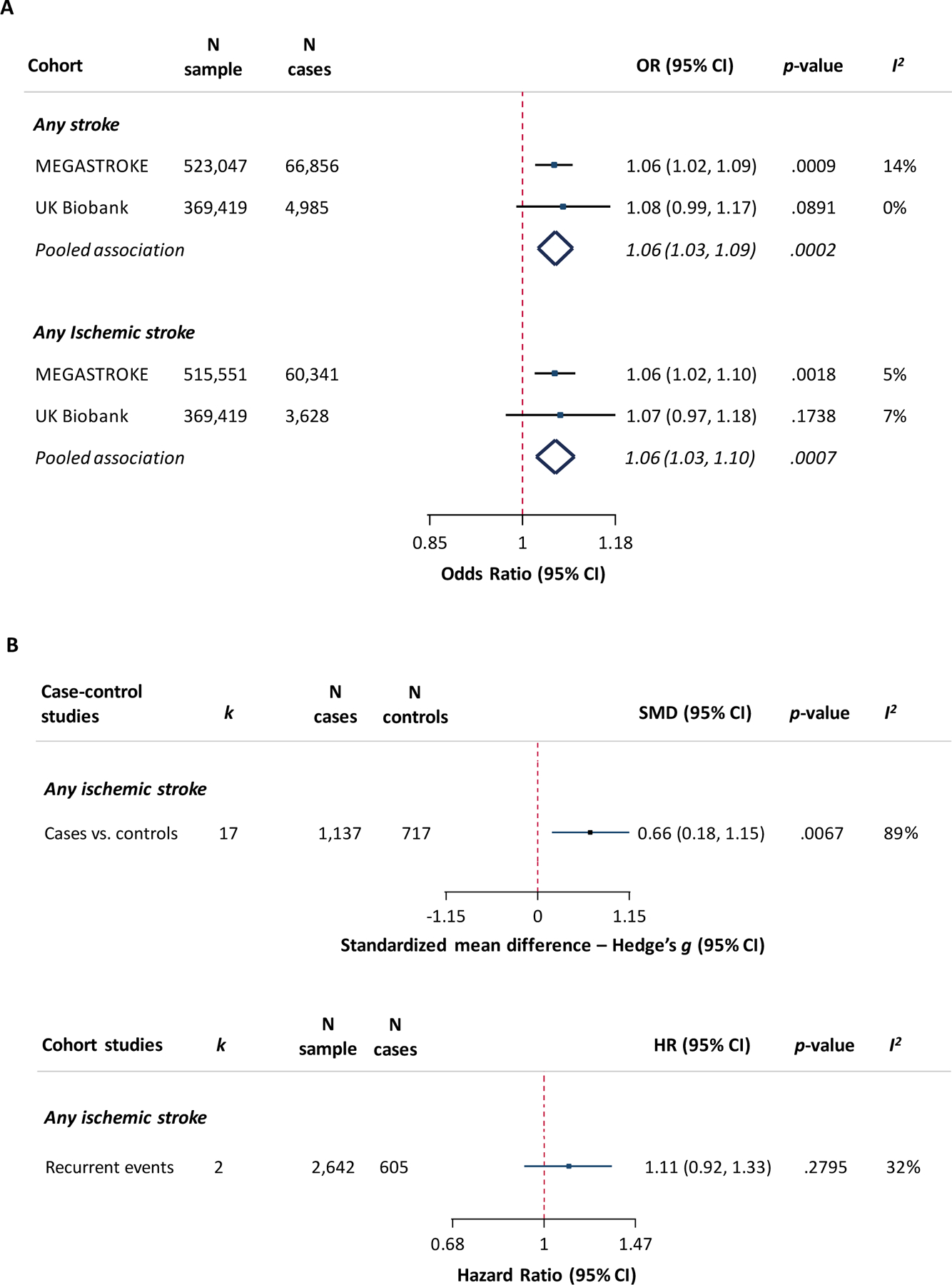
(A) MR-derived associations between genetically determined circulating MCP-1 levels (1-SD increase) and risk of any stroke and any ischemic stroke in MEGASTROKE, in UK Biobank, and a meta-analysis of both samples. (B) Meta-analysis-derived associations between circulating MCP-1 levels (1-SD increase) and risk of ischemic stroke in case-control and cohort studies. k refers to number of included studies. I2 in Figure 4A refers to heterogeneity in the Mendelian randomization analysis (inverse-variance weighted method) and in Figure 4B in the random-effects meta-analyses of observational studies.
CI, confidence interval; HR, hazard ratio; OR, odds ratio; SMD, standardized mean difference; SNP, single nucleotide polymorphism.
Circulating levels of MCP-1 and risk of stroke: meta-analysis of observational studies
Next, we compared the MR estimates with those derived from a meta-analysis of observational studies. Our search yielded 17 case-control studies of ischemic stroke patients and controls, two cohort studies on patients with a history of stroke or cardiovascular disease exploring the risk of recurrent ischemic stroke, and one case-cohort study of incident ischemic stroke in a community population (Supplemental Tables 5 and 6 and Supplemental Figure 6). Patients with any ischemic stroke were found to have significantly higher MCP-1 levels than controls in the case-control studies (Hedges’ g: 0.66, 95%CI: 0.18–1.15 [corresponding to a medium to strong effect size34]; 1137 cases, 717 controls; heterogeneity: I2=89%, p<0.001; Figure 4B and Supplemental Figure 7A). Studies on recurrent stroke (2,642 individuals, 605 events) yielded a HR of 1.11 (95%CI: 0.92–1.33) for 1 SD increase in MCP-1 levels (heterogeneity: I2=32%, p=0.23; Figure 4B and Supplemental Figure 7B), whereas the single study examining incident ischemic stroke (95 cases, 190 controls) reported a HR of 0.99 (95%CI: 0.68–1.45).
Genetically determined circulating levels of MCP-1 and etiologically related vascular outcomes
Figure 5 depicts the MR association between genetically determined MCP-1 levels and risk of CAD, MI and AF. Genetic predisposition to higher MCP-1 levels was associated with CAD (OR per 1-SD increase: 1.04, 95%CI: 1.00–1.08, p=0.04; 184,305 individuals, 60,801 cases) and MI (OR: 1.05, 95%CI: 1.01–1.09, p=0.02; 167,180 individuals, 43,676 cases). Given the association of MCP-1 with cardioembolic stroke, we further explored the relationship between genetically determined MCP-1 levels and risk of AF in MR analysis, but found no association (OR: 0.96, 95%CI: 0.91–1.01, p=0.09).
Figure 5. Mendelian randomization analysis for genetically determined circulating MCP-1 levels and etiologically related vascular outcomes.
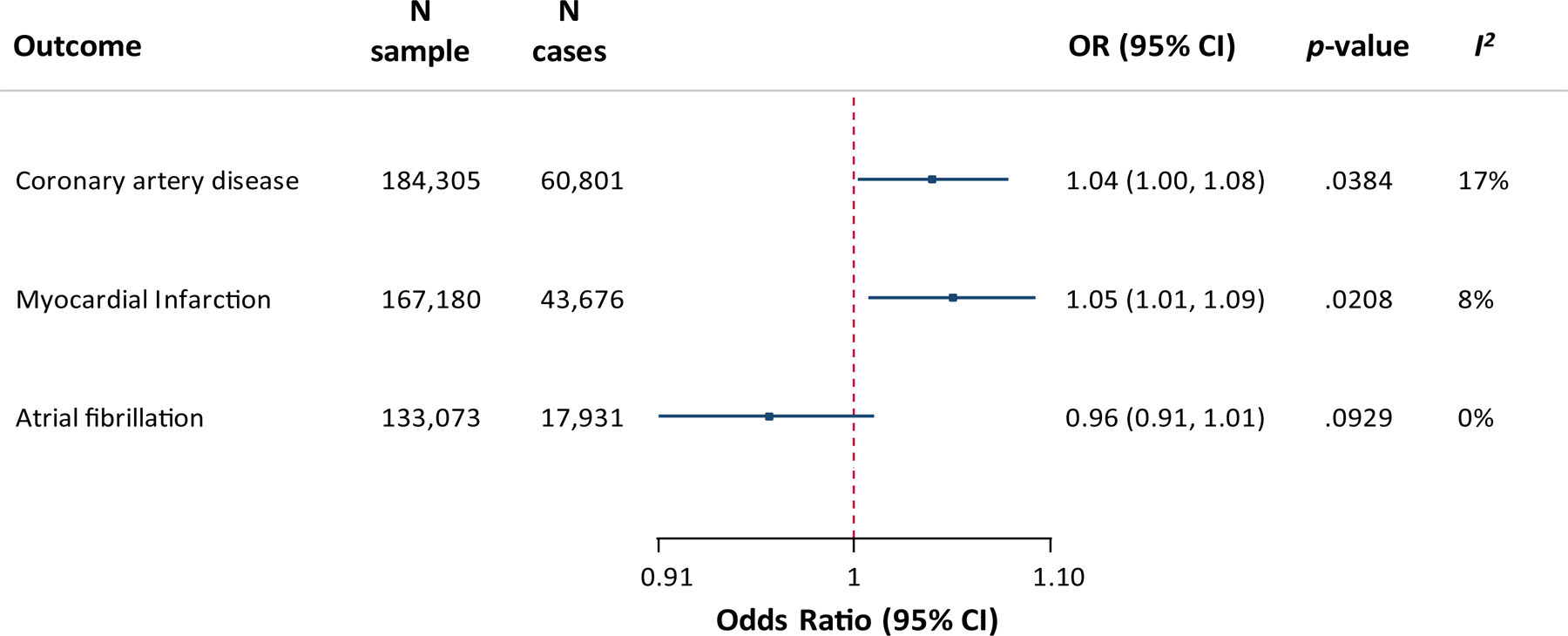
MR-derived associations between genetically determined circulating MCP-1 levels (1-SD increase) and risk of coronary artery disease, myocardial infarction, and atrial fibrillation. I2 refers to heterogeneity in the Mendelian randomization analysis (inverse-variance weighted method).
DISCUSSION
Exploring 41 cytokines in a two-sample MR approach involving the largest GWAS datasets available, we found that genetic predisposition to higher levels of MCP-1/CCL2 is associated with higher risk of any stroke, any ischemic stroke, large artery stroke, and cardioembolic stroke. The results were stable in alternative MR methods and sensitivity analyses and remained significant after adjustment for cardiovascular risk factors. Moreover, effect sizes for any stroke and any ischemic stroke were similar in the UK Biobank. We further found associations between genetic predisposition to higher MCP-1 levels and higher risk of CAD and MI as etiologically related outcomes. Collectively, our findings suggest that lifelong elevated circulating MCP-1 levels increase risk of stroke.
The directionality of the MR association between genetically determined levels of MCP-1 and risk of large artery stroke is consistent with experimental data showing a key role for this chemokine in atherogenesis and atheroprogression. Acting mainly through its receptor CCR2, MCP-1 is the prototypical CC family chemokine that is upregulated by chronic inflammatory conditions and attracts monocytes to the subendothelial space of the atherogenic arterial wall.35 Mice lacking MCP-136 or CCR237 are less susceptible to atherosclerosis and anti-MCP-1 gene therapy,38 MCP-1 competitors,39 and CCR2 antagonists40 reduce plaque size and inhibit plaque progression and destabilization in experimental atherosclerosis. Conversely, overexpression of MCP-1 leads to inflammation, accumulation of lipids, and smooth muscle cell proliferation in atherosclerotic plaques.41
We further found an MR association between genetic predisposition to higher MCP-1 levels and risk of cardioembolic stroke. Genetic predisposition to higher MCP-1 levels is associated with higher risk of coronary artery disease and myocardial infarction, which could promote the formation of left ventricular thrombus from myocardial damage thus resulting in cardioembolic stroke. Furthermore, MCP-1 has been reported to promote myocardial fibrosis,42 an established risk factor for AF.43 However, we found no association between the genetic instruments for MCP-1 and AF risk. Other investigators have found an association between circulating MCP-1 levels and the presence of atrial thrombi in patients with AF.44 Hence, it might be that MCP-1 increases the risk of cardioembolic stroke by promoting thrombus formation in patients with established AF. Alternative explanations for the association between circulating MCP-1 levels and cardioembolic stroke might include less frequent causes of cardioembolism such as valvular disease and misclassification of patients with multiple competing stroke etiologies including atherosclerosis.
In contrast, our analysis provides no evidence for an association of genetically determined MCP-1 levels with small vessel stroke even though the sample size was larger than for other stroke subtypes. In fact, we found none of the cytokines to be associated with small vessel stroke (all p>0.05, Figure 2). Overall, these observations agree with the notion that inflammatory processes are less important in small vessel disease than in large artery atherosclerosis although this has so far not been systematically examined.
The lack of a signal with intracerebral hemorrhage, and particularly deep intracerebral hemorrhage, which like small vessel stroke is attributed to small vessel disease,17 is in line with this result. However, this analysis was based on a rather small sample size. Also, following restriction of the analysis to SNPs in the vicinity of genes interacting with MCP-1, we identified a significant association between genetically determined MCP-1 levels and intracerebral hemorrhage. This difference in results might relate to exclusion of nonspecific instruments in the sensitivity analyses and should be explored further in larger samples.
Our meta-analysis of case-control studies revealed higher circulating MCP-1 levels in patients with ischemic stroke compared to healthy controls. Our systematic search identified only three prospective cohort studies, one on incident45 and two on recurrent stroke events,46, 47 none of which showed significant results. However, these studies had small sample sizes and a low number of events. Also, ischemic stroke subtypes were not considered, thus precluding meaningful comparisons with our MR results. Interestingly, observational cohort studies on CAD found higher MCP-1 levels to be associated with higher risk of incident48 and recurrent49 events consistent with the observed association with atherosclerotic stroke. Serial measurements of MCP-1 in large population-based cohorts with data on ischemic stroke subtypes would offer further insights into the relationship between MCP-1 and risk of stroke.
Targeting specific inflammatory cytokines might reduce vascular risk. The recent multicenter CANTOS trial showed that canakinumab, a monoclonal antibody against IL-1β, decreases the rate of recurrent cardiovascular events, including nonfatal myocardial infarction, nonfatal stroke and cardiovascular mortality, among patients with MI and elevated circulating CRP levels.8 Unfortunately, the original cytokine GWAS did not identify any genetic instruments for IL-1β circulating levels12 thus precluding a comparison of the MR results with the results of the CANTOS trial.8 The MCP-1/CCR2 pathway was targeted in a small phase II clinical trial in patients with risk factors for atherosclerosis and elevated circulating CRP levels. MLN1202, a humanized monoclonal antibody against CCR2 reduced CRP levels after 4 and 12 weeks.50 However, effects on clinical endpoints were not assessed50 and would need to be determined in a larger trial.
This study has several methodological strengths. We used the most recent and comprehensive dataset for cytokine levels and the largest available GWAS dataset for stroke and stroke subtypes. Results were confirmed through sensitivity analyses for pleiotropy including alternative MR methods, in sub-analyses on a biologically plausible protein-protein interaction network, and in analyses on etiologically related outcomes (CAD and MI).
Our study also has limitations. First, none of the SNPs used as instruments for MCP-1 were located in the vicinity of the MCP1 gene thus precluding analyses restricted to SNPs within this locus. Consequently, while we found no statistical evidence for pleiotropy, we cannot preclude nonspecific effects of the MCP-1 trans-acting instruments. Second, our instrument selection was based on a single discovery GWAS that adjusted for BMI. While the association remained consistent when using an unweighted allele score, we cannot exclude that the BMI adjustment led to collider bias during instrument selection. Third, we could not obtain reliable genetic instruments for 18 cytokines and several analyses for ischemic stroke subtypes were underpowered. Thus, we might have missed associations for several cytokines that have previously been implicated in vascular disease such as IL-1β, TNF-α and IL-6. Targeted studies incorporating further GWAS data on individual cytokines might reveal additional associations not captured by our approach. Fourth, genetic instruments were selected using an FDR-based approach, which might have weakened the instruments. However, the F-statistics were high and the results were in line with those derived when selecting instruments based on the genome-wide threshold (p <5×10−8). Finally, the UK Biobank analysis was rather underpowered and did not include stroke subtypes. Yet, the consistency of both the direction and magnitude of the associations between genetically determined MCP-1 and risk of any stroke and any ischemic stroke supports our results.
In conclusion, this study demonstrates that lifelong elevated circulating MCP-1 levels are associated with higher risk of stroke and particularly with the large artery and the cardioembolic subtypes. Future studies should explore in more depth whether targeting MCP-1 or its downstream effectors could be a meaningful strategy to reduce stroke risk.
Supplementary Material
CLINICAL PERSPECTIVE.
What is new?
Genetic predisposition to higher circulating levels of monocyte chemoattractant protein-1 (MCP-1/CCL2) was associated with higher risk of stroke
Associations were also found for etiologic stroke subtypes, specifically large artery stroke and cardioembolic stroke
Genetically determined levels of MCP-1 also associated with higher risk of the related phenotypes of coronary artery disease and myocardial infarction
What are the clinical implications?
Additional work is needed to determine whether targeting MCP-1 or its downstream effectors is a meaningful strategy for lowering stroke risk
Acknowledgements:
This research has been conducted using the UK Biobank Resource (UK Biobank application 2532, “UK Biobank stroke study: developing an in-depth understanding of the determinants of stroke and its subtypes”). We thank the following consortia for making data publicly available: CARDIoGRAMplusC4D Consortium, AFGen Consortium, GLGC Consortium, and DIAGRAM Consortium.
Funding sources: This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreements No 666881, “Small vessel diseases in a mechanistic perspective: Targets for Intervention” (SVDs@target; to M Dichgans) and No 667375, “Common mechanisms and pathways in stroke and Alzheimer’s disease” (CoSTREAM; to M Dichgans); the German Research Foundation (DFG) as part of the “Munich Cluster for Systems Neurology” (SyNergy; EXC 1010) and the Collaborative Research Center (CRC) 1123 (B3) (to M Dichgans); the Corona Foundation (to M Dichgans); the Foundation Leducq (Transatlantic Network of Excellence on the Pathogenesis of Small Vessel Disease of the Brain) (to M Dichgans); the e:Med program (“Systems medicine of myocardial infarction and stroke”; e:AtheroSysMed) (to M Dichgans); and the European Union’s Seventh Framework Programme (FP7/2007-2103) project “Exploitation of Genomic Variants Affecting Coronary Artery Disease and Stroke Risk for Therapeutic Intervention” (CVgenes@target; grant agreement number Health-F2-2013-601456; to M Dichgans). D Gill is funded by the Wellcome Trust. JC Hopewell is supported by a fellowship from the British Heart Foundation (FS/14/55/30806).
APPENDIX
Full author list of the MEGASTROKE consortium of the International Stroke Genetics Consortium (ISGC):
Rainer Malik 1, Ganesh Chauhan 2, Matthew Traylor 3, Muralidharan Sargurupremraj 4,5, Yukinori Okada 6,7,8, Aniket Mishra 4,5, Loes Rutten-Jacobs 3, Anne-Katrin Giese 9, Sander W van der Laan 10, Solveig Gretarsdottir 11, Christopher D Anderson 12,13,14,14, Michael Chong 15, Hieab HH Adams 16,17, Tetsuro Ago 18, Peter Almgren 19, Philippe Amouyel 20,21, Hakan Ay 22,13, Traci M Bartz 23, Oscar R Benavente 24, Steve Bevan 25, Giorgio B Boncoraglio 26, Robert D Brown, Jr. 27, Adam S Butterworth 28,29, Caty Carrera 30,31, Cara L Carty 32,33, Daniel I Chasman 34,35, Wei-Min Chen 36, John W Cole 37, Adolfo Correa 38, Ioana Cotlarciuc 39, Carlos Cruchaga 40,41, John Danesh 28,42,43,44, Paul IW de Bakker 45,46, Anita L DeStefano 47,48, Marcel den Hoed 49, Qing Duan 50, Stefan T Engelter 51,52, Guido J Falcone 53,54, Rebecca F Gottesman 55, Raji P Grewal 56, Vilmundur Gudnason 57,58, Stefan Gustafsson 59, Jeffrey Haessler 60, Tamara B Harris 61, Ahamad Hassan 62, Aki S Havulinna 63,64, Susan R Heckbert 65, Elizabeth G Holliday 66,67, George Howard 68, Fang-Chi Hsu 69, Hyacinth I Hyacinth 70, M Arfan Ikram 16, Erik Ingelsson 71,72, Marguerite R Irvin 73, Xueqiu Jian 74, Jordi Jiménez-Conde 75, Julie A Johnson 76,77, J Wouter Jukema 78, Masahiro Kanai 6,7,79, Keith L Keene 80,81, Brett M Kissela 82, Dawn O Kleindorfer 82, Charles Kooperberg 60, Michiaki Kubo 83, Leslie A Lange 84, Carl D Langefeld 85, Claudia Langenberg 86, Lenore J Launer 87, Jin-Moo Lee 88, Robin Lemmens 89,90, Didier Leys 91, Cathryn M Lewis 92,93, Wei-Yu Lin 28,94, Arne G Lindgren 95,96, Erik Lorentzen 97, Patrik K Magnusson 98, Jane Maguire 99, Ani Manichaikul 36, Patrick F McArdle 100, James F Meschia 101, Braxton D Mitchell 100,102, Thomas H Mosley 103,104, Michael A Nalls 105,106, Toshiharu Ninomiya 107, Martin J O’Donnell 15,108, Bruce M Psaty 109,110,111,112, Sara L Pulit 113,45, Kristiina Rannikmae 114,115, Alexander P Reiner 65,116, Kathryn M Rexrode 117, Kenneth Rice 118, Stephen S Rich 36, Paul M Ridker 34,35, Natalia S Rost 9,13, Peter M Rothwell 119, Jerome I Rotter 120,121, Tatjana Rundek 122, Ralph L Sacco 122, Saori Sakaue 7,123, Michele M Sale 124, Veikko Salomaa 63, Bishwa R Sapkota 125, Reinhold Schmidt 126, Carsten O Schmidt 127, Ulf Schminke 128, Pankaj Sharma 39, Agnieszka Slowik 129, Cathie LM Sudlow 114,115, Christian Tanislav 130, Turgut Tatlisumak 131,132, Kent D Taylor 120,121, Vincent NS Thijs 133,134, Gudmar Thorleifsson 11, Unnur Thorsteinsdottir 11, Steffen Tiedt 1, Stella Trompet 135, Christophe Tzourio 5,136,137, Cornelia M van Duijn 138,139, Matthew Walters 140, Nicholas J Wareham 86, Sylvia Wassertheil-Smoller 141, James G Wilson 142, Kerri L Wiggins 109, Qiong Yang 47, Salim Yusuf 15, Najaf Amin 16, Hugo S Aparicio 185,48, Donna K Arnett 186, John Attia 187, Alexa S Beiser 47,48, Claudine Berr 188, Julie E Buring 34,35, Mariana Bustamante 189, Valeria Caso 190, Yu-Ching Cheng 191, Seung Hoan Choi 192,48, Ayesha Chowhan 185,48, Natalia Cullell 31, Jean-François Dartigues 193,194, Hossein Delavaran 95,96, Pilar Delgado 195, Marcus Dorr 196,197, Gunnar Engström 19, Ian Ford 198, Wander S Gurpreet 199, Anders Hamsten 200,201, Laura Heitsch 202, Atsushi Hozawa 203, Laura Ibanez 204, Andreea Ilinca 95,96, Martin Ingelsson 205, Motoki Iwasaki 206, Rebecca D Jackson 207, Katarina Jood 208, Pekka Jousilahti 63, Sara Kaffashian 4,5, Lalit Kalra 209, Masahiro Kamouchi 210, Takanari Kitazono 211, Olafur Kjartansson 212, Manja Kloss 213, Peter J Koudstaal 214, Jerzy Krupinski 215, Daniel L Labovitz 216, Cathy C Laurie 118, Christopher R Levi 217, Linxin Li 218, Lars Lind 219, Cecilia M Lindgren 220,221, Vasileios Lioutas 222,48, Yong Mei Liu 223, Oscar L Lopez 224, Hirata Makoto 225, Nicolas Martinez-Majander 172, Koichi Matsuda 225, Naoko Minegishi 203, Joan Montaner 226, Andrew P Morris 227,228, Elena Muino 31, Martina Müller-Nurasyid 229,230,231, Bo Norrving 95,96, Soichi Ogishima 203, Eugenio A Parati 232, Leema Reddy Peddareddygari 56, Nancy L Pedersen 98,233, Joanna Pera 129, Markus Perola 63,234, Alessandro Pezzini 235, Silvana Pileggi 236, Raquel Rabionet 237, Iolanda Riba-Llena 30, Marta Ribases 238, Jose R Romero 185,48, Jaume Roquer 239,240, Anthony G Rudd 241,242, Antti-Pekka Sarin 243,244, Ralhan Sarju 199, Chloe Sarnowski 47,48, Makoto Sasaki 245, Claudia L Satizabal 185,48, Mamoru Satoh 245, Naveed Sattar 246, Norie Sawada 206, Gerli Sibolt 172, Ásgeir Sigurdsson 247, Albert Smith 248, Kenji Sobue 245, Carolina Soriano-Tárraga 240, Tara Stanne 249, O Colin Stine 250, David J Stott 251, Konstantin Strauch 229,252, Takako Takai 203, Hideo Tanaka 253,254, Kozo Tanno 245, Alexander Teumer 255, Liisa Tomppo 172, Nuria P Torres-Aguila 31, Emmanuel Touze 256,257, Shoichiro Tsugane 206, Andre G Uitterlinden 258, Einar M Valdimarsson 259, Sven J van der Lee 16, Henry Völzke 255, Kenji Wakai 253, David Weir 260, Stephen R Williams 261, Charles DA Wolfe 241,242, Quenna Wong 118, Huichun Xu 191, Taiki Yamaji 206, Dharambir K Sanghera 125,169,170, Olle Melander 19, Christina Jern 171, Daniel Strbian 172,173, Israel Fernandez-Cadenas 31,30, W T Longstreth, Jr 174,65, Arndt Rolfs 175, Jun Hata 107, Daniel Woo 82, Jonathan Rosand 12,13,14, Guillaume Pare 15, Jemma C Hopewell 176, Danish Saleheen 177, Kari Stefansson 11,178, Bradford B Worrall 179, Steven J Kittner 37, Sudha Seshadri 180,48, Myriam Fornage 74,181, Hugh S Markus 3, Joanna MM Howson 28, Yoichiro Kamatani 6,182, Stephanie Debette 4,5, Martin Dichgans 1,183,184
1 Institute for Stroke and Dementia Research (ISD), University Hospital, LMU Munich, Munich, Germany
2 Centre for Brain Research, Indian Institute of Science, Bangalore, India
3 Stroke Research Group, Division of Clinical Neurosciences, University of Cambridge, UK
4 INSERM U1219 Bordeaux Population Health Research Center, Bordeaux, France
5 University of Bordeaux, Bordeaux, France
6 Laboratory for Statistical Analysis, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan
7 Department of Statistical Genetics, Osaka University Graduate School of Medicine, Osaka, Japan
8 Laboratory of Statistical Immunology, Immunology Frontier Research Center (WPI-IFReC), Osaka University, Suita, Japan.
9 Department of Neurology, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA
10 Laboratory of Experimental Cardiology, Division of Heart and Lungs, University Medical Center Utrecht, University of Utrecht, Utrecht,Netherlands
11 deCODE genetics/AMGEN inc, Reykjavik, Iceland
12 Center for Genomic Medicine, Massachusetts General Hospital (MGH), Boston, MA, USA
13 J. Philip Kistler Stroke Research Center, Department of Neurology, MGH, Boston, MA, USA
14 Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA
15 Population Health Research Institute, McMaster University, Hamilton, Canada
16 Department of Epidemiology, Erasmus University Medical Center, Rotterdam, Netherlands
17 Department of Radiology and Nuclear Medicine, Erasmus University Medical Center, Rotterdam, Netherlands
18 Department of Medicine and Clinical Science, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan
19 Department of Clinical Sciences, Lund University, Malmo, Sweden
20 Univ. Lille, Inserm, Institut Pasteur de Lille, LabEx DISTALZ-UMR1167, Risk factors and molecular determinants of aging-related diseases, F-59000 Lille, France
21 Centre Hosp. Univ Lille, Epidemiology and Public Health Department, F-59000 Lille, France
22 AA Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA
23 Cardiovascular Health Research Unit, Departments of Biostatistics and Medicine, University of Washington, Seattle, WA, USA
24 Division of Neurology, Faculty of Medicine, Brain Research Center, University of British Columbia, Vancouver, Canada
25 School of Life Science, University of Lincoln, Lincoln, UK
26 Department of Cerebrovascular Diseases, Fondazione IRCCS Istituto Neurologico “Carlo Besta”, Milano, Italy
27 Department of Neurology, Mayo Clinic Rochester, Rochester, MN, USA
28 MRC/BHF Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK
29 The National Institute for Health Research Blood and Transplant Research Unit in Donor Health and Genomics, University of Cambridge, UK
30 Neurovascular Research Laboratory, Vall d’Hebron Institut of Research, Neurology and Medicine Departments-Universitat Autonoma de Barcelona, Vall d’Hebrón Hospital, Barcelona, Spain
31 Stroke Pharmacogenomics and Genetics, Fundacio Docència i Recerca MutuaTerrassa, Terrassa, Spain
32 Children’s Research Institute, Children’s National Medical Center, Washington, DC, USA
33 Center for Translational Science, George Washington University, Washington, DC, USA
34 Division of Preventive Medicine, Brigham and Women’s Hospital, Boston, MA, USA
35 Harvard Medical School, Boston, MA, USA
36 Center for Public Health Genomics, Department of Public Health Sciences, University of Virginia, Charlottesville, VA, USA
37 Department of Neurology, University of Maryland School of Medicine and Baltimore VAMC, Baltimore, MD, USA
38 Departments of Medicine, Pediatrics and Population Health Science, University of Mississippi Medical Center, Jackson, MS, USA
39 Institute of Cardiovascular Research, Royal Holloway University of London, UK & Ashford and St Peters Hospital, Surrey UK
40 Department of Psychiatry,The Hope Center Program on Protein Aggregation and Neurodegeneration (HPAN),Washington University, School of Medicine, St. Louis, MO, USA
41 Department of Developmental Biology, Washington University School of Medicine, St. Louis, MO, USA
42 NIHR Blood and Transplant Research Unit in Donor Health and Genomics, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK
43 Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK
44 British Heart Foundation, Cambridge Centre of Excellence, Department of Medicine, University of Cambridge, Cambridge, UK
45 Department of Medical Genetics, University Medical Center Utrecht, Utrecht, Netherlands
46 Department of Epidemiology, Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht, Netherlands
47 Boston University School of Public Health, Boston, MA, USA
48 Framingham Heart Study, Framingham, MA, USA
49 Department of Immunology, Genetics and Pathology and Science for Life Laboratory, Uppsala University, Uppsala, Sweden
50 Department of Genetics, University of North Carolina, Chapel Hill, NC, USA
51 Department of Neurology and Stroke Center, Basel University Hospital, Switzerland
52 Neurorehabilitation Unit, University and University Center for Medicine of Aging and Rehabilitation Basel, Felix Platter Hospital, Basel, Switzerland
53 Department of Neurology, Yale University School of Medicine, New Haven, CT, USA
54 Program in Medical and Population Genetics, The Broad Institute of Harvard and MIT, Cambridge, MA, USA
55 Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD, USA
56 Neuroscience Institute, SF Medical Center, Trenton, NJ, USA
57 Icelandic Heart Association Research Institute, Kopavogur, Iceland
58 University of Iceland, Faculty of Medicine, Reykjavik, Iceland
59 Department of Medical Sciences, Molecular Epidemiology and Science for Life Laboratory, Uppsala University, Uppsala, Sweden
60 Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA
61 Laboratory of Epidemiology and Population Science, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA
62 Department of Neurology, Leeds General Infirmary, Leeds Teaching Hospitals NHS Trust, Leeds, UK
63 National Institute for Health and Welfare, Helsinki, Finland
64 FIMM - Institute for Molecular Medicine Finland, Helsinki, Finland
65 Department of Epidemiology, University of Washington, Seattle, WA, USA
66 Public Health Stream, Hunter Medical Research Institute, New Lambton, Australia
67 Faculty of Health and Medicine, University of Newcastle, Newcastle, Australia
68 School of Public Health, University of Alabama at Birmingham, Birmingham, AL, USA
69 Department of Biostatistical Sciences, Wake Forest School of Medicine, Winston-Salem, NC, USA
70 Aflac Cancer and Blood Disorder Center, Department of Pediatrics, Emory University School of Medicine, Atlanta, GA, USA
71 Department of Medicine, Division of Cardiovascular Medicine, Stanford University School of Medicine, CA, USA
72 Department of Medical Sciences, Molecular Epidemiology and Science for Life Laboratory, Uppsala University, Uppsala, Sweden
73 Epidemiology, School of Public Health, University of Alabama at Birmingham, USA
74 Brown Foundation Institute of Molecular Medicine, University of Texas Health Science Center at Houston, Houston, TX, USA
75 Neurovascular Research Group (NEUVAS), Neurology Department, Institut Hospital del Mar d’Investigacio Medica, Universitat Autonoma de Barcelona, Barcelona, Spain
76 Department of Pharmacotherapy and Translational Research and Center for Pharmacogenomics, University of Florida, College of Pharmacy, Gainesville, FL, USA
77 Division of Cardiovascular Medicine, College of Medicine, University of Florida, Gainesville, FL, USA
78 Department of Cardiology, Leiden University Medical Center, Leiden, the Netherlands
79 Program in Bioinformatics and Integrative Genomics, Harvard Medical School, Boston, MA, USA
80 Department of Biology, East Carolina University, Greenville, NC, USA
81 Center for Health Disparities, East Carolina University, Greenville, NC, USA
82 University of Cincinnati College of Medicine, Cincinnati, OH, USA
83 RIKEN Center for Integrative Medical Sciences, Yokohama, Japan
84 Department of Medicine, University of Colorado Denver, Anschutz Medical Campus, Aurora, CO, USA
85 Center for Public Health Genomics and Department of Biostatistical Sciences, Wake Forest School of Medicine, Winston-Salem, NC, USA
86 MRC Epidemiology Unit, University of Cambridge School of Clinical Medicine, Institute of Metabolic Science, Cambridge Biomedical Campus, Cambridge, UK
87 Intramural Research Program, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA
88 Department of Neurology, Radiology, and Biomedical Engineering, Washington University School of Medicine, St. Louis, MO, USA
89 KU Leuven - University of Leuven, Department of Neurosciences, Experimental Neurology, Leuven, Belgium
90 VIB Center for Brain & Disease Research, University Hospitals Leuven, Department of Neurology, Leuven, Belgium
91 Univ.-Lille, INSERM U 1171. CHU Lille. Lille, France
92 Department of Medical and Molecular Genetics, King’s College London, London, UK
93 SGDP Centre, Institute of Psychiatry, Psychology & Neuroscience, King’s College London, London, UK
94 Northern Institute for Cancer Research, Paul O’Gorman Building, Newcastle University, Newcastle, UK
95 Department of Clinical Sciences Lund, Neurology, Lund University, Lund, Sweden
96 Department of Neurology and Rehabilitation Medicine, Skane University Hospital, Lund, Sweden
97 Bioinformatics Core Facility, University of Gothenburg, Gothenburg, Sweden
98 Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden
99 University of Technology Sydney, Faculty of Health, Ultimo, Australia
100 Department of Medicine, University of Maryland School of Medicine, MD, USA
101 Department of Neurology, Mayo Clinic, Jacksonville, FL, USA
102 Geriatrics Research and Education Clinical Center, Baltimore Veterans Administration Medical Center, Baltimore, MD, USA
103 Division of Geriatrics, School of Medicine, University of Mississippi Medical Center, Jackson, MS, USA
104 Memory Impairment and Neurodegenerative Dementia Center, University of Mississippi Medical Center, Jackson, MS, USA
105 Laboratory of Neurogenetics, National Institute on Aging, National institutes of Health, Bethesda, MD, USA
106 Data Tecnica International, Glen Echo MD, USA
107 Department of Epidemiology and Public Health, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan
108 Clinical Research Facility, Department of Medicine, NUI Galway, Galway, Ireland
109 Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA, USA
110 Department of Epidemiology, University of Washington, Seattle, WA
111 Department of Health Services, University of Washington, Seattle, WA, USA
112 Kaiser Permanente Washington Health Research Institute, Seattle, WA, USA
113 Brain Center Rudolf Magnus, Department of Neurology, University Medical Center Utrecht, Utrecht, The Netherlands
114 Usher Institute of Population Health Sciences and Informatics, University of Edinburgh, Edinburgh, UK
115 Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, UK
116 Fred Hutchinson Cancer Research Center, University of Washington, Seattle, WA, USA
117 Department of Medicine, Brigham and Women’s Hospital, Boston, MA, USA
118 Department of Biostatistics, University of Washington, Seattle, WA, USA
119 Nuffield Department of Clinical Neurosciences, University of Oxford, UK
120 Institute for Translational Genomics and Population Sciences, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA, USA
121 Division of Genomic Outcomes, Department of Pediatrics, Harbor-UCLA Medical Center, Torrance, CA, USA
122 Department of Neurology, Miller School of Medicine, University of Miami, Miami, FL, USA
123 Department of Allergy and Rheumatology, Graduate School of Medicine, the University of Tokyo, Tokyo, Japan
124 Center for Public Health Genomics, University of Virginia, Charlottesville, VA, USA
125 Department of Pediatrics, College of Medicine, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA
126 Department of Neurology, Medical University of Graz, Graz, Austria
127 University Medicine Greifswald, Institute for Community Medicine, SHIP-KEF, Greifswald, Germany
128 University Medicine Greifswald, Department of Neurology, Greifswald, Germany
129 Department of Neurology, Jagiellonian University, Krakow, Poland
130 Department of Neurology, Justus Liebig University, Giessen, Germany
131 Department of Clinical Neurosciences/Neurology, Institute of Neuroscience and Physiology, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden
132 Sahlgrenska University Hospital, Gothenburg, Sweden
133 Stroke Division, Florey Institute of Neuroscience and Mental Health, University of Melbourne, Heidelberg, Australia
134 Austin Health, Department of Neurology, Heidelberg, Australia
135 Department of Internal Medicine, Section Gerontology and Geriatrics, Leiden University Medical Center, Leiden, the Netherlands
136 INSERM U1219, Bordeaux, France
137 Department of Public Health, Bordeaux University Hospital, Bordeaux, France
138 Genetic Epidemiology Unit, Department of Epidemiology, Erasmus University Medical Center Rotterdam, Netherlands
139 Center for Medical Systems Biology, Leiden, Netherlands
140 School of Medicine, Dentistry and Nursing at the University of Glasgow, Glasgow, UK
141 Department of Epidemiology and Population Health, Albert Einstein College of Medicine, NY, USA
142 Department of Physiology and Biophysics, University of Mississippi Medical Center, Jackson, MS, USA
143 A full list of members and affiliations appears in the Supplementary Note
144 Department of Human Genetics, McGill University, Montreal, Canada
145 Department of Pathophysiology, Institute of Biomedicine and Translation Medicine, University of Tartu, Tartu, Estonia
146 Department of Cardiac Surgery, Tartu University Hospital, Tartu, Estonia
147 Clinical Gene Networks AB,Stockholm, Sweden
148 Department of Genetics and Genomic Sciences, The Icahn Institute for Genomics and Multiscale Biology Icahn School of Medicine at Mount Sinai, New York, NY , USA
149 Department of Pathophysiology, Institute of Biomedicine and Translation Medicine, University of Tartu, Biomeedikum, Tartu, Estonia
150 Integrated Cardio Metabolic Centre, Department of Medicine, Karolinska Institutet, Karolinska Universitetssjukhuset, Huddinge, Sweden.
151 Clinical Gene Networks AB, Stockholm, Sweden
152 Sorbonne Universites, UPMC Univ. Paris 06, INSERM, UMR_S 1166, Team Genomics & Pathophysiology of Cardiovascular Diseases, Paris, France
153 ICAN Institute for Cardiometabolism and Nutrition, Paris, France
154 Department of Biomedical Engineering, University of Virginia, Charlottesville, VA, USA
155 Group Health Research Institute, Group Health Cooperative, Seattle, WA, USA
156 Seattle Epidemiologic Research and Information Center, VA Office of Research and Development, Seattle, WA, USA
157 Cardiovascular Research Center, Massachusetts General Hospital, Boston, MA, USA
158 Department of Medical Research, B^rum Hospital, Vestre Viken Hospital Trust, Gjettum, Norway
159 Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore
160 National Heart and Lung Institute, Imperial College London, London, UK
161 Department of Gene Diagnostics and Therapeutics, Research Institute, National Center for Global Health and Medicine, Tokyo, Japan
162 Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA, USA
163 Department of Cardiology,University Medical Center Groningen, University of Groningen, Netherlands
164 MRC-PHE Centre for Environment and Health, School of Public Health, Department of Epidemiology and Biostatistics, Imperial College London, London, UK
165 Department of Epidemiology and Biostatistics, Imperial College London, London, UK
166 Department of Cardiology, Ealing Hospital NHS Trust, Southall, UK
167 National Heart, Lung and Blood Research Institute, Division of Intramural Research, Population Sciences Branch, Framingham, MA, USA
168 A full list of members and affiliations appears at the end of the manuscript
169 Department of Phamaceutical Sciences, Collge of Pharmacy, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA
170 Oklahoma Center for Neuroscience, Oklahoma City, OK, USA
171 Department of Pathology and Genetics, Institute of Biomedicine, The Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden
172 Department of Neurology, Helsinki University Hospital, Helsinki, Finland
173 Clinical Neurosciences, Neurology, University of Helsinki, Helsinki, Finland
174 Department of Neurology, University of Washington, Seattle, WA, USA
175 Albrecht Kossel Institute, University Clinic of Rostock, Rostock, Germany
176 Clinical Trial Service Unit and Epidemiological Studies Unit, Nuffield Department of Population Health, University of Oxford, Oxford, UK
177 Department of Genetics, Perelman School of Medicine, University of Pennsylvania, PA, USA
178 Faculty of Medicine, University of Iceland, Reykjavik, Iceland
179 Departments of Neurology and Public Health Sciences, University of Virginia School of Medicine, Charlottesville, VA, USA
180 Department of Neurology, Boston University School of Medicine, Boston, MA, USA
181 Human Genetics Center, University of Texas Health Science Center at Houston, Houston, TX, USA
182 Center for Genomic Medicine, Kyoto University Graduate School of Medicine, Kyoto, Japan
183 Munich Cluster for Systems Neurology (SyNergy), Munich, Germany
184 German Center for Neurodegenerative Diseases (DZNE), Munich, Germany
185 Boston University School of Medicine, Boston, MA, USA
186 University of Kentucky College of Public Health, Lexington, KY, USA
187 University of Newcastle and Hunter Medical Research Institute, New Lambton, Australia
188 Univ. Montpellier, Inserm, U1061, Montpellier, France
189 Centre for Research in Environmental Epidemiology, Barcelona, Spain
190 Department of Neurology, Università degli Studi di Perugia, Umbria, Italy
191 Department of Medicine, University of Maryland School of Medicine, Baltimore, MD, USA
192 Broad Institute, Cambridge, MA, USA
193 Univ. Bordeaux, Inserm, Bordeaux Population Health Research Center, UMR 1219, Bordeaux, France
194 Bordeaux University Hospital, Department of Neurology, Memory Clinic, Bordeaux, France
195 Neurovascular Research Laboratory. Vall d’Hebron Institut of Research, Neurology and Medicine Departments-Universitat Autonoma de Barcelona. Vall d’Hebrón Hospital, Barcelona, Spain
196 University Medicine Greifswald, Department of Internal Medicine B, Greifswald, Germany
197 DZHK, Greifswald, Germany
198 Robertson Center for Biostatistics, University of Glasgow, Glasgow, UK
199 Hero DMC Heart Institute, Dayanand Medical College & Hospital, Ludhiana, India
200 Atherosclerosis Research Unit, Department of Medicine Solna, Karolinska Institutet, Stockholm, Sweden
201 Karolinska Institutet, Stockholm, Sweden
202 Division of Emergency Medicine, and Department of Neurology, Washington University School of Medicine, St. Louis, MO, USA
203 Tohoku Medical Megabank Organization, Sendai, Japan
204 Department of Psychiatry, Washington University School of Medicine, St. Louis, MO, USA
205 Department of Public Health and Caring Sciences / Geriatrics, Uppsala University, Uppsala, Sweden
206 Epidemiology and Prevention Group, Center for Public Health Sciences, National Cancer Center, Tokyo, Japan
207 Department of Internal Medicine and the Center for Clinical and Translational Science, The Ohio State University, Columbus, OH, USA
208 Institute of Neuroscience and Physiology, the Sahlgrenska Academy at University of Gothenburg, Goteborg, Sweden
209 Department of Basic and Clinical Neurosciences, King’s College London, London, UK
210 Department of Health Care Administration and Management, Graduate School of Medical Sciences, Kyushu University, Japan
211 Department of Medicine and Clinical Science, Graduate School of Medical Sciences, Kyushu University, Japan
212 Landspitali National University Hospital, Departments of Neurology & Radiology, Reykjavik, Iceland
213 Department of Neurology, Heidelberg University Hospital, Germany
214 Department of Neurology, Erasmus University Medical Center
215 Hospital Universitari Mutua Terrassa, Terrassa (Barcelona), Spain
216 Albert Einstein College of Medicine, Montefiore Medical Center, New York, NY, USA
217 John Hunter Hospital, Hunter Medical Research Institute and University of Newcastle, Newcastle, NSW, Australia
218 Centre for Prevention of Stroke and Dementia, Nuffield Department of Clinical Neurosciences, University of Oxford, UK
219 Department of Medical Sciences, Uppsala University, Uppsala, Sweden
220 Genetic and Genomic Epidemiology Unit, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK
221 The Wellcome Trust Centre for Human Genetics, Oxford, UK
222 Beth Israel Deaconess Medical Center, Boston, MA, USA
223 Wake Forest School of Medicine, Wake Forest, NC, USA
224 Department of Neurology, University of Pittsburgh, Pittsburgh, PA, USA
225 BioBank Japan, Laboratory of Clinical Sequencing, Department of Computational biology and medical Sciences, Graduate school of Frontier Sciences, The University of Tokyo, Tokyo, Japan
226 Neurovascular Research Laboratory, Vall d’Hebron Institut of Research, Neurology and Medicine Departments-Universitat Autonoma de Barcelona. Vall d’Hebron Hospital, Barcelona, Spain
227 Department of Biostatistics, University of Liverpool, Liverpool, UK
228 Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK
229 Institute of Genetic Epidemiology, Helmholtz Zentrum München - German Research Center for Environmental Health, Neuherberg, Germany
230 Department of Medicine I, Ludwig-Maximilians-Universität, Munich, Germany
231 DZHK (German Centre for Cardiovascular Research), partner site Munich Heart Alliance, Munich, Germany
232 Department of Cerebrovascular Diseases, Fondazione IRCCS Istituto Neurologico “Carlo Besta”, Milano, Italy
233 Karolinska Institutet, MEB, Stockholm, Sweden
234 University of Tartu, Estonian Genome Center, Tartu, Estonia, Tartu, Estonia
235 Department of Clinical and Experimental Sciences, Neurology Clinic, University of Brescia, Italy
236 Translational Genomics Unit, Department of Oncology, IRCCS Istituto di Ricerche Farmacologiche Mario Negri, Milano, Italy
237 Department of Genetics, Microbiology and Statistics, University of Barcelona, Barcelona, Spain
238 Psychiatric Genetics Unit, Group of Psychiatry, Mental Health and Addictions, Vall d’Hebron Research Institute (VHIR), Universitat Autònoma de Barcelona, Biomedical Network Research Centre on Mental Health (CIBERSAM), Barcelona, Spain
239 Department of Neurology, IMIM-Hospital del Mar, and Universitat Autònoma de Barcelona, Spain
240 IMIM (Hospital del Mar Medical Research Institute), Barcelona, Spain
241 National Institute for Health Research Comprehensive Biomedical Research Centre, Guy’s & St. Thomas’ NHS Foundation Trust and King’s College London, London, UK
242 Division of Health and Social Care Research, King’s College London, London, UK
243 FIMM-Institute for Molecular Medicine Finland, Helsinki, Finland
244 THL-National Institute for Health and Welfare, Helsinki, Finland
245 Iwate Tohoku Medical Megabank Organization, Iwate Medical University,
Iwate, Japan
246 BHF Glasgow Cardiovascular Research Centre, Faculty of Medicine, Glasgow, UK
247 deCODE Genetics/Amgen, Inc., Reykjavik, Iceland
248 Icelandic Heart Association, Reykjavik, Iceland
249 Institute of Biomedicine, the Sahlgrenska Academy at University of Gothenburg, Goteborg, Sweden
250 Department of Epidemiology, University of Maryland School of Medicine, Baltimore, MD, USA
251 Institute of Cardiovascular and Medical Sciences, Faculty of Medicine, University of Glasgow, Glasgow, UK
252 Chair of Genetic Epidemiology, IBE, Faculty of Medicine, LMU Munich, Germany
253 Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Japan
254 Department of Epidemiology, Nagoya University Graduate School of Medicine, Nagoya, Japan
255 University Medicine Greifswald, Institute for Community Medicine, SHIP-KEF, Greifswald, Germany
256 Department of Neurology, Caen University Hospital, Caen, France
257 University of Caen Normandy, Caen, France
258 Department of Internal Medicine, Erasmus University Medical Center, Rotterdam, Netherlands
259 Landspitali University Hospital, Reykjavik, Iceland
260 Survey Research Center, University of Michigan, Ann Arbor, MI, USA
261 University of Virginia Department of Neurology, Charlottesville, VA, USA
Footnotes
Disclosures: None.
REFERENCES
- 1.DALYs GBD and Collaborators H. Global, regional, and national disability-adjusted life-years (DALYs) for 315 diseases and injuries and healthy life expectancy (HALE), 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1603–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mortality GBD and Causes of Death C. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1459–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feigin VL, Norrving B and Mensah GA. Global Burden of Stroke. Circ Res. 2017;120:439–448. [DOI] [PubMed] [Google Scholar]
- 4.Esenwa CC and Elkind MS. Inflammatory risk factors, biomarkers and associated therapy in ischaemic stroke. Nat Rev Neurol. 2016;12:594–604. [DOI] [PubMed] [Google Scholar]
- 5.Libby P, Ridker PM and Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. [DOI] [PubMed] [Google Scholar]
- 6.Rouhl RP, Damoiseaux JG, Lodder J, Theunissen RO, Knottnerus IL, Staals J, Henskens LH, Kroon AA, de Leeuw PW, Tervaert JW and van Oostenbrugge RJ. Vascular inflammation in cerebral small vessel disease. NeurobiolAging. 2012;33:1800–1806. [DOI] [PubMed] [Google Scholar]
- 7.Ridker PM. Targeting inflammatory pathways for the treatment of cardiovascular disease. Eur Heart J. 2014;35:540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, Kastelein JJP, Cornel JH, Pais P, Pella D, Genest J, Cifkova R, Lorenzatti A, Forster T, Kobalava Z, Vida-Simiti L, Flather M, Shimokawa H, Ogawa H, Dellborg M, Rossi PRF, Troquay RPT, Libby P, Glynn RJ and Group CT. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N Engl J Med. 2017;377:1119–1131. [DOI] [PubMed] [Google Scholar]
- 9.Smith GD and Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. [DOI] [PubMed] [Google Scholar]
- 10.Kelly PJ, Murphy S, Coveney S, Purroy F, Lemmens R, Tsivgoulis G and Price C. Anti-inflammatory approaches to ischaemic stroke prevention. J Neurol Neurosurg Psychiatry. 2018;89:211–218. [DOI] [PubMed] [Google Scholar]
- 11.Holmes MV, Ala-Korpela M and Smith GD. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol. 2017;14:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahola-Olli AV, Wurtz P, Havulinna AS, Aalto K, Pitkanen N, Lehtimaki T, Kahonen M, Lyytikainen LP, Raitoharju E, Seppala I, Sarin AP, Ripatti S, Palotie A, Perola M, Viikari JS, Jalkanen S, Maksimow M, Salomaa V, Salmi M, Kettunen J and Raitakari OT. Genome-wide Association Study Identifies 27 Loci Influencing Concentrations of Circulating Cytokines and Growth Factors. Am J Hum Genet. 2017;100:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese AK, van der Laan SW, Gretarsdottir S, Anderson CD, Chong M, Adams HHH, Ago T, Almgren P, Amouyel P, Ay H, Bartz TM, Benavente OR, Bevan S, Boncoraglio GB, Brown RD Jr., Butterworth AS, Carrera C, Carty CL, Chasman DI, Chen WM, Cole JW, Correa A, Cotlarciuc I, Cruchaga C, Danesh J, de Bakker PIW, DeStefano AL, den Hoed M, Duan Q, Engelter ST, Falcone GJ, Gottesman RF, Grewal RP, Gudnason V, Gustafsson S, Haessler J, Harris TB, Hassan A, Havulinna AS, Heckbert SR, Holliday EG, Howard G, Hsu FC, Hyacinth HI, Ikram MA, Ingelsson E, Irvin MR, Jian X, Jimenez-Conde J, Johnson JA, Jukema JW, Kanai M, Keene KL, Kissela BM, Kleindorfer DO, Kooperberg C, Kubo M, Lange LA, Langefeld CD, Langenberg C, Launer LJ, Lee JM, Lemmens R, Leys D, Lewis CM, Lin WY, Lindgren AG, Lorentzen E, Magnusson PK, Maguire J, Manichaikul A, McArdle PF, Meschia JF, Mitchell BD, Mosley TH, Nalls MA, Ninomiya T, O’Donnell MJ, Psaty BM, Pulit SL, Rannikmae K, Reiner AP, Rexrode KM, Rice K, Rich SS, Ridker PM, Rost NS, Rothwell PM, Rotter JI, Rundek T, Sacco RL, Sakaue S, Sale MM, Salomaa V, Sapkota BR, Schmidt R, Schmidt CO, Schminke U, Sharma P, Slowik A, Sudlow CLM, Tanislav C, Tatlisumak T, Taylor KD, Thijs VNS, Thorleifsson G, Thorsteinsdottir U, Tiedt S, Trompet S, Tzourio C, van Duijn CM, Walters M, Wareham NJ, Wassertheil-Smoller S, Wilson JG, Wiggins KL, Yang Q, Yusuf S, Bis JC, Pastinen T, Ruusalepp A, Schadt EE, Koplev S, Bjorkegren JLM, Codoni V, Civelek M, Smith NL, Tregouet DA, Christophersen IE, Roselli C, Lubitz SA, Ellinor PT, Tai ES, Kooner JS, Kato N, He J, van der Harst P, Elliott P, Chambers JC, Takeuchi F, Johnson AD, Sanghera DK, Melander O, Jern C, Strbian D, Fernandez-Cadenas I, Longstreth WT Jr., Rolfs A, Hata J, Woo D, Rosand J, Pare G, Hopewell JC, Saleheen D, Stefansson K, Worrall BB, Kittner SJ, Seshadri S, Fornage M, Markus HS, Howson JMM, Kamatani Y, Debette S, Dichgans M, Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese AK, van der Laan SW, Gretarsdottir S, Anderson CD, Chong M, Adams HHH, Ago T, Almgren P, Amouyel P, Ay H, Bartz TM, Benavente OR, Bevan S, Boncoraglio GB, Brown RD Jr., Butterworth AS, Carrera C, Carty CL, Chasman DI, Chen WM, Cole JW, Correa A, Cotlarciuc I, Cruchaga C, Danesh J, de Bakker PIW, DeStefano AL, Hoed MD, Duan Q, Engelter ST, Falcone GJ, Gottesman RF, Grewal RP, Gudnason V, Gustafsson S, Haessler J, Harris TB, Hassan A, Havulinna AS, Heckbert SR, Holliday EG, Howard G, Hsu FC, Hyacinth HI, Ikram MA, Ingelsson E, Irvin MR, Jian X, Jimenez-Conde J, Johnson JA, Jukema JW, Kanai M, Keene KL, Kissela BM, Kleindorfer DO, Kooperberg C, Kubo M, Lange LA, Langefeld CD, Langenberg C, Launer LJ, Lee JM, Lemmens R, Leys D, Lewis CM, Lin WY, Lindgren AG, Lorentzen E, Magnusson PK, Maguire J, Manichaikul A, McArdle PF, Meschia JF, Mitchell BD, Mosley TH, Nalls MA, Ninomiya T, O’Donnell MJ, Psaty BM, Pulit SL, Rannikmae K, Reiner AP, Rexrode KM, Rice K, Rich SS, Ridker PM, Rost NS, Rothwell PM, Rotter JI, Rundek T, Sacco RL, Sakaue S, Sale MM, Salomaa V, Sapkota BR, Schmidt R, Schmidt CO, Schminke U, Sharma P, Slowik A, Sudlow CLM, Tanislav C, Tatlisumak T, Taylor KD, Thijs VNS, Thorleifsson G, Thorsteinsdottir U, Tiedt S, Trompet S, Tzourio C, van Duijn CM, Walters M, Wareham NJ, Wassertheil-Smoller S, Wilson JG, Wiggins KL, Yang Q, Yusuf S, Amin N, Aparicio HS, Arnett DK, Attia J, Beiser AS, Berr C, Buring JE, Bustamante M, Caso V, Cheng YC, Choi SH, Chowhan A, Cullell N, Dartigues JF, Delavaran H, Delgado P, Dorr M, Engstrom G, Ford I, Gurpreet WS, Hamsten A, Heitsch L, Hozawa A, Ibanez L, Ilinca A, Ingelsson M, Iwasaki M, Jackson RD, Jood K, Jousilahti P, Kaffashian S, Kalra L, Kamouchi M, Kitazono T, Kjartansson O, Kloss M, Koudstaal PJ, Krupinski J, Labovitz DL, Laurie CC, Levi CR, Li L, Lind L, Lindgren CM, Lioutas V, Liu YM, Lopez OL, Makoto H, Martinez-Majander N, Matsuda K, Minegishi N, Montaner J, Morris AP, Muino E, Muller-Nurasyid M, Norrving B, Ogishima S, Parati EA, Peddareddygari LR, Pedersen NL, Pera J, Perola M, Pezzini A, Pileggi S, Rabionet R, Riba-Llena I, Ribases M, Romero JR, Roquer J, Rudd AG, Sarin AP, Sarju R, Sarnowski C, Sasaki M, Satizabal CL, Satoh M, Sattar N, Sawada N, Sibolt G, Sigurdsson A, Smith A, Sobue K, Soriano-Tarraga C, Stanne T, Stine OC, Stott DJ, Strauch K, Takai T, Tanaka H, Tanno K, Teumer A, Tomppo L, Torres-Aguila NP, Touze E, Tsugane S, Uitterlinden AG, Valdimarsson EM, van der Lee SJ, Volzke H, Wakai K, Weir D, Williams SR, Wolfe CDA, Wong Q, Xu H, Yamaji T, Sanghera DK, Melander O, Jern C, Strbian D, Fernandez-Cadenas I, Longstreth WT Jr., Rolfs A, Hata J, Woo D, Rosand J, Pare G, Hopewell JC, Saleheen D, Stefansson K, Worrall BB, Kittner SJ, Seshadri S, Fornage M, Markus HS, Howson JMM, Kamatani Y, Debette S, Dichgans M, Consortium AF, Cohorts for H, Aging Research in Genomic Epidemiology C, International Genomics of Blood Pressure C, Consortium I, Starnet, BioBank Japan Cooperative Hospital G, Consortium C, Consortium E-C, Consortium EP-I, International Stroke Genetics C, Consortium M, Neurology Working Group of the CC, Network NSG, Study UKYLD, Consortium M and Consortium M. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, Giannakopoulou O, Jiang T, Hamby SE, Di Angelantonio E, Assimes TL, Bottinger EP, Chambers JC, Clarke R, Palmer CNA, Cubbon RM, Ellinor P, Ermel R, Evangelou E, Franks PW, Grace C, Gu D, Hingorani AD, Howson JMM, Ingelsson E, Kastrati A, Kessler T, Kyriakou T, Lehtimaki T, Lu X, Lu Y, Marz W, McPherson R, Metspalu A, Pujades-Rodriguez M, Ruusalepp A, Schadt EE, Schmidt AF, Sweeting MJ, Zalloua PA, AlGhalayini K, Keavney BD, Kooner JS, Loos RJF, Patel RS, Rutter MK, Tomaszewski M, Tzoulaki I, Zeggini E, Erdmann J, Dedoussis G, Bjorkegren JLM, Consortium E-C, CardioGramplusC4D, group UKBCCCw, Schunkert H, Farrall, Danesh J, Samani NJ, Watkins H and Deloukas P. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. [DOI] [PubMed] [Google Scholar]
- 15.Holmes MV, Asselbergs FW, Palmer TM, Drenos F, Lanktree MB, Nelson CP, Dale CE, Padmanabhan S, Finan C, Swerdlow DI, Tragante V, van Iperen EP, Sivapalaratnam S, Shah S, Elbers CC, Shah T, Engmann J, Giambartolomei C, White J, Zabaneh D, Sofat R, McLachlan S, consortium U, Doevendans PA, Balmforth AJ, Hall AS, North KE, Almoguera B, Hoogeveen RC, Cushman M, Fornage M, Patel SR, Redline S, Siscovick DS, Tsai MY, Karczewski KJ, Hofker MH, Verschuren WM, Bots ML, van der Schouw YT, Melander O, Dominiczak AF, Morris R, Ben-Shlomo Y, Price J, Kumari M, Baumert J, Peters A, Thorand B, Koenig W, Gaunt TR, Humphries SE, Clarke R, Watkins H, Farrall M, Wilson JG, Rich SS, de Bakker PI, Lange LA, Davey Smith G, Reiner AP, Talmud PJ, Kivimaki M, Lawlor DA, Dudbridge F, Samani NJ, Keating BJ, Hingorani AD and Casas JP. Mendelian randomization of blood lipids for coronary heart disease. Eur Heart J. 2015;36:539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams HP Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL and Marsh EE 3rd. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41. [DOI] [PubMed] [Google Scholar]
- 17.Woo D, Falcone GJ, Devan WJ, Brown WM, Biffi A, Howard TD, Anderson CD, Brouwers HB, Valant V, Battey TW, Radmanesh F, Raffeld MR, Baedorf-Kassis S, Deka R, Woo JG, Martin LJ, Haverbusch M, Moomaw CJ, Sun G, Broderick JP, Flaherty ML, Martini SR, Kleindorfer DO, Kissela B, Comeau ME, Jagiella JM, Schmidt H, Freudenberger P, Pichler A, Enzinger C, Hansen BM, Norrving B, Jimenez-Conde J, Giralt-Steinhauer E, Elosua R, Cuadrado-Godia E, Soriano C, Roquer J, Kraft P, Ayres AM, Schwab K, McCauley JL, Pera J, Urbanik A, Rost NS, Goldstein JN, Viswanathan A, Stogerer EM, Tirschwell DL, Selim M, Brown DL, Silliman SL, Worrall BB, Meschia JF, Kidwell CS, Montaner J, Fernandez-Cadenas I, Delgado P, Malik R, Dichgans M, Greenberg SM, Rothwell PM, Lindgren A, Slowik A, Schmidt R, Langefeld CD, Rosand J and International Stroke Genetics C. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet. 2014;94:511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierce BL, Ahsan H and Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40:740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol. 2014;43:922–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer TM, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, Timpson NJ, Davey Smith G and Sterne JA. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. 2012;21:223–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, Webb TR, Zeng L, Dehghan A, Alver M, Armasu SM, Auro K, Bjonnes A, Chasman DI, Chen S, Ford I, Franceschini N, Gieger C, Grace C, Gustafsson S, Huang J, Hwang SJ, Kim YK, Kleber ME, Lau KW, Lu X, Lu Y, Lyytikainen LP, Mihailov E, Morrison AC, Pervjakova N, Qu L, Rose LM, Salfati E, Saxena R, Scholz M, Smith AV, Tikkanen E, Uitterlinden A, Yang X, Zhang W, Zhao W, de Andrade M, de Vries PS, van Zuydam NR, Anand SS, Bertram L, Beutner F, Dedoussis G, Frossard P, Gauguier D, Goodall AH, Gottesman O, Haber M, Han BG, Huang J, Jalilzadeh S, Kessler T, Konig IR, Lannfelt L, Lieb W, Lind L, Lindgren CM, Lokki ML, Magnusson PK, Mallick NH, Mehra N, Meitinger T, Memon FU, Morris AP, Nieminen MS, Pedersen NL, Peters A, Rallidis LS, Rasheed A, Samuel M, Shah SH, Sinisalo J, Stirrups KE, Trompet S, Wang L, Zaman KS, Ardissino D, Boerwinkle E, Borecki IB, Bottinger EP, Buring JE, Chambers JC, Collins R, Cupples LA, Danesh J, Demuth I, Elosua R, Epstein SE, Esko T, Feitosa MF, Franco OH, Franzosi MG, Granger CB, Gu D, Gudnason V, Hall AS, Hamsten A, Harris TB, Hazen SL, Hengstenberg C, Hofman A, Ingelsson E, Iribarren C, Jukema JW, Karhunen PJ, Kim BJ, Kooner JS, Kullo IJ, Lehtimaki T, Loos RJF, Melander O, Metspalu A, Marz W, Palmer CN, Perola M, Quertermous T, Rader DJ, Ridker PM, Ripatti S, Roberts R, Salomaa V, Sanghera DK, Schwartz SM, Seedorf U, Stewart AF, Stott DJ, Thiery J, Zalloua PA, O’Donnell CJ, Reilly MP, Assimes TL, Thompson JR, Erdmann J, Clarke R, Watkins H, Kathiresan S, McPherson R, Deloukas P, Schunkert H, Samani NJ and Farrall M. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christophersen IE, Rienstra M, Roselli C, Yin X, Geelhoed B, Barnard J, Lin H, Arking DE, Smith AV, Albert CM, Chaffin M, Tucker NR, Li M, Klarin D, Bihlmeyer NA, Low SK, Weeke PE, Muller-Nurasyid M, Smith JG, Brody JA, Niemeijer MN, Dorr M, Trompet S, Huffman J, Gustafsson S, Schurmann C, Kleber ME, Lyytikainen LP, Seppala I, Malik R, Horimoto A, Perez M, Sinisalo J, Aeschbacher S, Theriault S, Yao J, Radmanesh F, Weiss S, Teumer A, Choi SH, Weng LC, Clauss S, Deo R, Rader DJ, Shah SH, Sun A, Hopewell JC, Debette S, Chauhan G, Yang Q, Worrall BB, Pare G, Kamatani Y, Hagemeijer YP, Verweij N, Siland JE, Kubo M, Smith JD, Van Wagoner DR, Bis JC, Perz S, Psaty BM, Ridker PM, Magnani JW, Harris TB, Launer LJ, Shoemaker MB, Padmanabhan S, Haessler J, Bartz TM, Waldenberger M, Lichtner P, Arendt M, Krieger JE, Kahonen M, Risch L, Mansur AJ, Peters A, Smith BH, Lind L, Scott SA, Lu Y, Bottinger EB, Hernesniemi J, Lindgren CM, Wong JA, Huang J, Eskola M, Morris AP, Ford I, Reiner AP, Delgado G, Chen LY, Chen YI, Sandhu RK, Li M, Boerwinkle E, Eisele L, Lannfelt L, Rost N, Anderson CD, Taylor KD, Campbell A, Magnusson PK, Porteous D, Hocking LJ, Vlachopoulou E, Pedersen NL, Nikus K, Orho-Melander M, Hamsten A, Heeringa J, Denny JC, Kriebel J, Darbar D, Newton-Cheh C, Shaffer C, Macfarlane PW, Heilmann-Heimbach S, Almgren P, Huang PL, Sotoodehnia N, Soliman EZ, Uitterlinden AG, Hofman A, Franco OH, Volker U, Jockel KH, Sinner MF, Lin HJ, Guo X, ISGC MCot, Neurology Working Group of the CC, Dichgans M, Ingelsson E, Kooperberg C, Melander O, Loos RJF, Laurikka J, Conen D, Rosand J, van der Harst P, Lokki ML, Kathiresan S, Pereira A, Jukema JW, Hayward C, Rotter JI, Marz W, Lehtimaki, Stricker BH, Chung MK, Felix SB, Gudnason V, Alonso A, Roden DM, Kaab S, Chasman DI, Heckbert SR, Benjamin EJ, Tanaka T, Lunetta KL, Lubitz SA, Ellinor PT and Consortium AF. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat Genet. 2017;49:946–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess S, Butterworth A and Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greco MF, Minelli C, Sheehan NA and Thompson JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. 2015;34:2926–2940. [DOI] [PubMed] [Google Scholar]
- 25.Bowden J, Davey Smith G and Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowden J, Davey Smith G, Haycock PC and Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verbanck M, Chen CY, Neale B and Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgess S and Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkila K, Hypponen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikainen LP, Magnusson PKE, Mangino M, Mihailov E, Montasser ME, Muller-Nurasyid M, Nolte IM, O’Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney ASF, Doring A, Elliott P, Epstein SE, Ingi Eyjolfsson G, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJP, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimaki T, Lin SY, Lindstrom J, Loos RJF, Mach F, McArdle WL, Meisinger C, Mitchell BD, Muller G, Nagaraja R, Narisu N, Nieminen TVM, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancakova A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YI, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrieres J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Jarvelin MR, Jula A, Kahonen M, Kaprio J, Kesaniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, Marz W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njolstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PEH, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BHR, Ordovas JM, Boerwinkle E, Palmer CNA, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR and Global Lipids Genetics C. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J, Lindgren CM, Muller-Nurasyid M, Pechlivanis S, Rayner NW, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S, Johnson AD, Dimas AS, Loos RJ, Vedantam S, Chen H, Florez JC, Fox C, Liu CT, Rybin D, Couper DJ, Kao WH, Li M, Cornelis MC, Kraft P, Sun Q, van Dam RM, Stringham HM, Chines PS, Fischer K, Fontanillas P, Holmen OL, Hunt SE, Jackson AU, Kong A, Lawrence R, Meyer J, Perry JR, Platou CG, Potter S, Rehnberg E, Robertson N, Sivapalaratnam S, Stancakova A, Stirrups K, Thorleifsson G, Tikkanen E, Wood AR, Almgren P, Atalay M, Benediktsson R, Bonnycastle LL, Burtt N, Carey J, Charpentier G, Crenshaw AT, Doney AS, Dorkhan M, Edkins S, Emilsson V, Eury E, Forsen T, Gertow K, Gigante B, Grant GB, Groves CJ, Guiducci C, Herder C, Hreidarsson AB, Hui J, James A, Jonsson A, Rathmann W, Klopp N, Kravic J, Krjutskov K, Langford C, Leander K, Lindholm E, Lobbens S, Mannisto S, Mirza G, Muhleisen TW, Musk B, Parkin M, Rallidis L, Saramies J, Sennblad B, Shah S, Sigurethsson G, Silveira A, Steinbach G, Thorand B, Trakalo J, Veglia F, Wennauer R, Winckler W, Zabaneh D, Campbell H, van Duijn C, Uitterlinden AG, Hofman A, Sijbrands E, Abecasis GR, Owen KR, Zeggini E, Trip MD, Forouhi NG, Syvanen AC, Eriksson JG, Peltonen L, Nothen MM, Balkau B, Palmer CN, Lyssenko V, Tuomi T, Isomaa B, Hunter DJ, Qi L, Wellcome Trust Case Control C, Meta-Analyses of G, Insulin-related traits Consortium I, Genetic Investigation of ATC, Asian Genetic Epidemiology Network-Type 2 Diabetes C, South Asian Type 2 Diabetes C, Shuldiner AR, Roden M, Barroso I, Wilsgaard T, Beilby J, Hovingh K, Price JF, Wilson JF, Rauramaa R, Lakka TA, Lind L, Dedoussis G, Njolstad I, Pedersen NL, Khaw KT, Wareham NJ, Keinanen-Kiukaanniemi SM, Saaristo TE, Korpi-Hyovalti E, Saltevo J, Laakso M, Kuusisto J, Metspalu A, Collins FS, Mohlke KL, Bergman RN, Tuomilehto J, Boehm BO, Gieger C, Hveem K, Cauchi S, Froguel P, Baldassarre D, Tremoli E, Humphries SE, Saleheen D, Danesh J, Ingelsson E, Ripatti S, Salomaa V, Erbel R, Jockel KH, Moebus S, Peters A, Illig T, de Faire U, Hamsten A, Morris AD, Donnelly PJ, Frayling TM, Hattersley AT, Boerwinkle E, Melander O, Kathiresan S, Nilsson PM, Deloukas P, Thorsteinsdottir U, Groop LC, Stefansson K, Hu F, Pankow JS, Dupuis J, Meigs JB, Altshuler D, Boehnke M, McCarthy MI, Replication DIG and Meta-analysis C. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ and von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ioannidis JP, Ntzani EE, Trikalinos TA and Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. [DOI] [PubMed] [Google Scholar]
- 33.Charoen P, Nitsch D, Engmann J, Shah T, White J, Zabaneh D, Jefferis B, Wannamethee G, Whincup P, Mulick Cassidy A, Gaunt T, Day I, McLachlan S, Price J, Kumari M, Kivimaki M, Brunner E, Langenberg C, Ben-Shlomo Y, Hingorani A, Whittaker J, Pablo Casas J, Dudbridge F and Consortium U. Mendelian Randomisation study of the influence of eGFR on coronary heart disease. Sci Rep. 2016;6:28514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen J. Statistical power analysis for the behavioral sciences. 2nd ed. Hillsdale, NJ: Erlbaum; 1988. [Google Scholar]
- 35.Lin J, Kakkar V and Lu X. Impact of MCP-1 in atherosclerosis. Curr Pharm Des. 2014;20:4580–4588. [DOI] [PubMed] [Google Scholar]
- 36.Gu L, Okada Y, Clinton SK, Gerard C, Sukhova GK, Libby P and Rollins BJ. Absence of monocyte chemoattractant protein-1 reduces atherosclerosis in low density lipoprotein receptor-deficient mice. Mol Cell. 1998;2:275–281. [DOI] [PubMed] [Google Scholar]
- 37.Boring L, Gosling J, Cleary M and Charo IF. Decreased lesion formation in CCR2−/−mice reveals a role for chemokines in the initiation of atherosclerosis. Nature. 1998;394:894–897. [DOI] [PubMed] [Google Scholar]
- 38.Inoue S, Egashira K, Ni W, Kitamoto S, Usui M, Otani K, Ishibashi M, Hiasa K, Nishida K and Takeshita A. Anti-monocyte chemoattractant protein-1 gene therapy limits progression and destabilization of established atherosclerosis in apolipoprotein E-knockout mice. Circulation. 2002;106:2700–2706. [DOI] [PubMed] [Google Scholar]
- 39.Liehn EA, Piccinini AM, Koenen RR, Soehnlein O, Adage T, Fatu R, Curaj A, Popescu A, Zernecke A, Kungl AJ and Weber C. A new monocyte chemotactic protein-1/chemokine CC motif ligand-2 competitor limiting neointima formation and myocardial ischemia/reperfusion injury in mice. J Am Coll Cardiol. 2010;56:1847–1857. [DOI] [PubMed] [Google Scholar]
- 40.Bot I, Ortiz Zacarias NV, de Witte WE, de Vries H, van Santbrink PJ, van der Velden D, Kroner MJ, van der Berg DJ, Stamos D, de Lange EC, Kuiper J, AP IJ and Heitman LH. A novel CCR2 antagonist inhibits atherogenesis in apoE deficient mice by achieving high receptor occupancy. SciRep. 2017;7:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aiello RJ, Bourassa PA, Lindsey S, Weng W, Natoli E, Rollins BJ and Milos PM. Monocyte chemoattractant protein-1 accelerates atherosclerosis in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol. 1999;19:1518–1525. [DOI] [PubMed] [Google Scholar]
- 42.Biernacka A and Frangogiannis NG. Aging and Cardiac Fibrosis. Aging Dis. 2011;2:158–173. [PMC free article] [PubMed] [Google Scholar]
- 43.Dzeshka MS, Lip GY, Snezhitskiy V and Shantsila E. Cardiac Fibrosis in Patients With Atrial Fibrillation: Mechanisms and Clinical Implications. J Am Coll Cardiol. 2015;66:943–959. [DOI] [PubMed] [Google Scholar]
- 44.Hammwohner M, Ittenson A, Dierkes J, Bukowska A, Klein HU, Lendeckel U and Goette A. Platelet expression of CD40/CD40 ligand and its relation to inflammatory markers and adhesion molecules in patients with atrial fibrillation. Exp Biol Med (Maywood). 2007;232:581–589. [PubMed] [Google Scholar]
- 45.Canoui-Poitrine F, Luc G, Mallat Z, Machez E, Bingham A, Ferrieres J, Ruidavets JB, Montaye M, Yarnell J, Haas B, Arveiler D, Morange P, Kee F, Evans A, Amouyel P, Ducimetiere P, Empana JP and Group PS. Systemic chemokine levels, coronary heart disease, and ischemic stroke events: the PRIME study. Neurology. 2011;77:1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganz P, Amarenco P, Goldstein LB, Sillesen H, Bao W, Preston GM, Welch KMA and Committee SS. Association of Osteopontin, Neopterin, and Myeloperoxidase With Stroke Risk in Patients With Prior Stroke or Transient Ischemic Attacks: Results of an Analysis of 13 Biomarkers From the Stroke Prevention by Aggressive Reduction in Cholesterol Levels Trial. Stroke. 2017;48:3223–3231. [DOI] [PubMed] [Google Scholar]
- 47.Haim M, Tanne D, Boyko V, Reshef T, Goldbourt U, Battler A, Mekori YA and Behar S. Monocyte chemoattractant protein-1 and recurrent cardiovascular events in patients with stable coronary heart disease. Clin Cardiol. 2005;28:31–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoogeveen RC, Morrison A, Boerwinkle E, Miles JS, Rhodes CE, Sharrett AR and Ballantyne CM. Plasma MCP-1 level and risk for peripheral arterial disease and incident coronary heart disease: Atherosclerosis Risk in Communities study. Atherosclerosis. 2005;183:301–307. [DOI] [PubMed] [Google Scholar]
- 49.de Lemos JA, Morrow DA, Sabatine MS, Murphy SA, Gibson CM, Antman EM, McCabe CH, Cannon CP and Braunwald E. Association between plasma levels of monocyte chemoattractant protein-1 and long-term clinical outcomes in patients with acute coronary syndromes. Circulation. 2003;107:690–695. [DOI] [PubMed] [Google Scholar]
- 50.Gilbert J, Lekstrom-Himes J, Donaldson D, Lee Y, Hu M, Xu J, Wyant T, Davidson M and Group MLNS. Effect of CC chemokine receptor 2 CCR2 blockade on serum C-reactive protein in individuals at atherosclerotic risk and with a single nucleotide polymorphism of the monocyte chemoattractant protein-1 promoter region. Am J Cardiol. 2011;107:906–911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


