Figure 6. Epitope Mapping of HVEM.
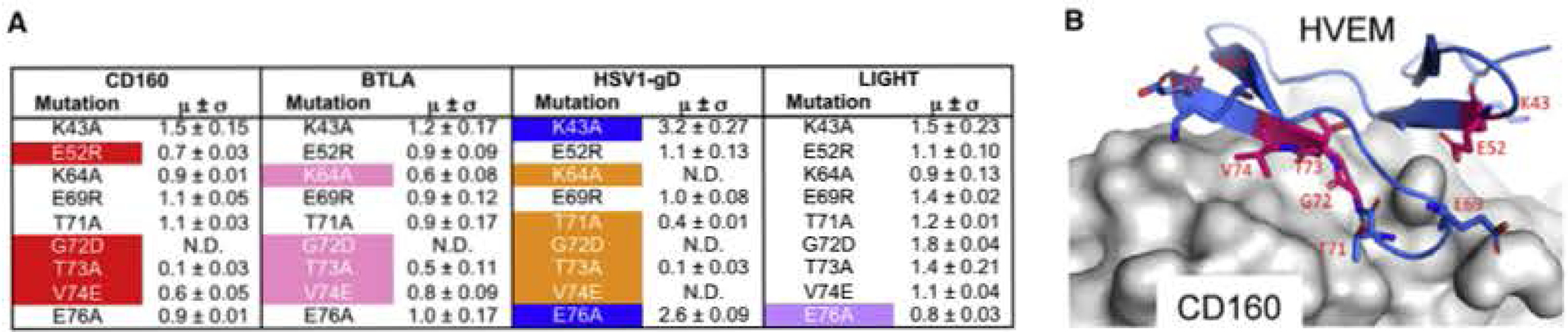
(A) Epitope mapping results of human HVEM, The relative binding affinities are represented by the values when normalized to the wild-type HVEM binding affinity (standard deviations are results of triplicates). N.D. stands for no detectable binding. The residues are categorized as sensitive residues when more than 20% binding affinity was lost. These affected residue’s are highlighted by color. Two residues with more than 2-fold increased affinities are colored blue.
(B) Structural representation of the CD160:HVEM epitope mapping results. The HVEM residues subjected to epitope mapping are shown as sticks. The sensitive residues with more than 20% compromised binding are highlighted by purple color. CD160 is shown as gray surface.
