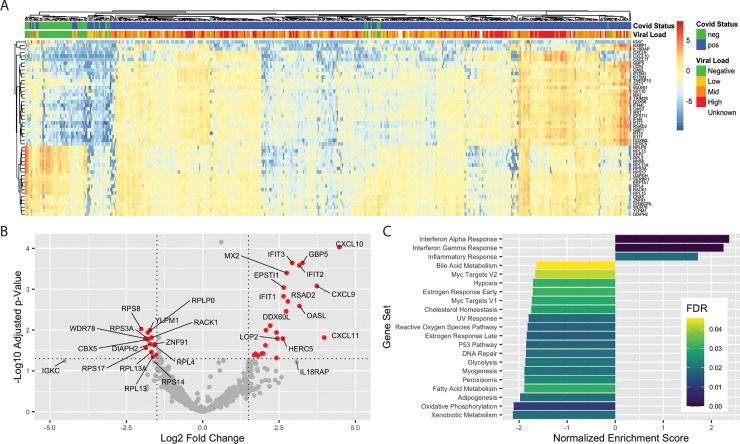
Fig 1. DE genes in SARS-CoV-2 NP swabs.
(A) Clustering of samples based on 50 genes with the lowest adjusted p-value. Log2 fold changes relative to gene mean are displayed by color. (B) Volcano plot of 15 most up-regulated and 15 most down-regulated genes in SARS-CoV-2 positive samples relative to negative by log2 fold change. Red color indicates genes with log2 fold change > |1.5| and adjusted p < 0.05. (C). Significant (FDR < 0.05) pathways affected by SARS-CoV-2 infection identified by GSEA. Raw data available in the GEO Repository, accession GSE152075. DE, differentially expressed; FDR, false discovery rate; GEO, Gene Expression Omnibus; GSEA, Gene Set Enrichment Analysis; NP, nasopharyngeal; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.