Figure 1.
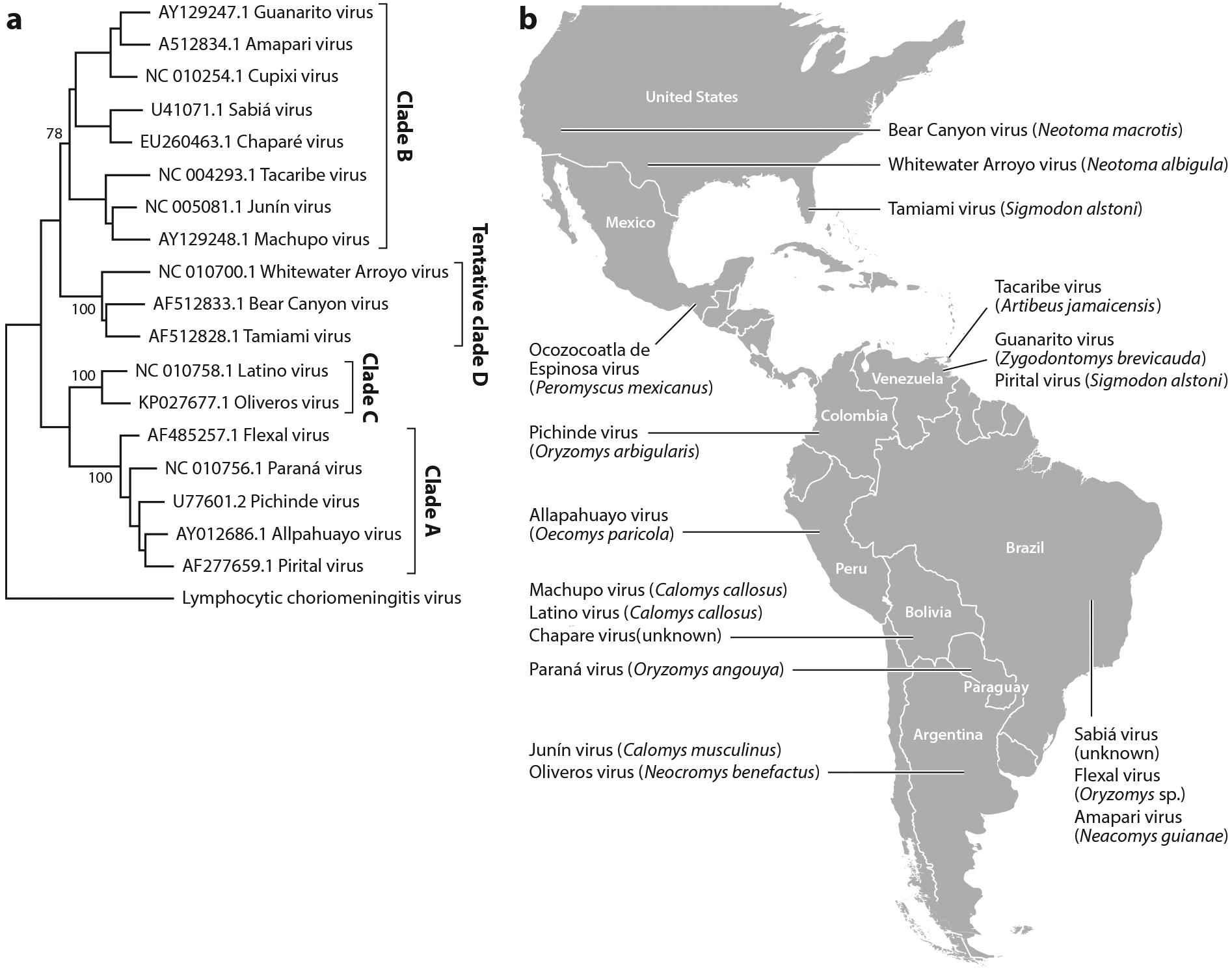
New World arenavirus taxonomy and geographic distribution. (a) Cladogram inferred by using the maximum-likelihood method with the Kimura two-parameter and gamma distribution substitution model. Phylogenetic reconstruction was performed using the full-length glycoprotein precursor (GPC) gene. Bootstrap values are shown for the main nodes. The GenBank accession number and origin of each sequence are denoted, as are the clade names. A strain of lymphocytic choriomeningitis virus was used as outgroup.
(b) Map of the Americas showing the distribution of the New World arenaviruses. Virus names and most frequent host reservoirs are indicated in the boxes.
