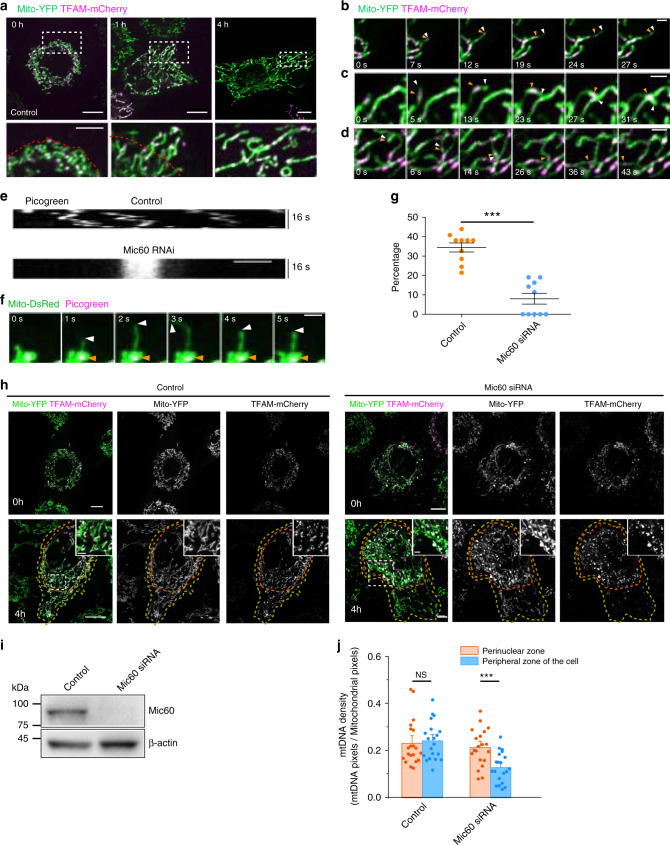
Fig. 5. MDT contributes to nucleoids distribution in peripheral mitochondrial network.
a Confocal images of KIF5B−/− NRK cells expressing mito-YFP, TFAM-mCherry, and TET-ON-KIF5B and treated with 0.5 µg/ml tetracycline. Higher magnifications of the white dashed boxes are in the lower panels. Red dashed lines indicate the mitochondrial network boundary before tubulation. b–d KIF5B−/− NRK cells expressing mito-YFP, TFAM-mCherry, and TET-ON-KIF5B were treated with 0.5 µg/ml tetracycline for 1 h (b, c) and 2 h (d). White arrows indicate a dynamic tubule. Orange arrowheads indicate nucleoids. e Kymographs of mtDNA labeled by DNA dye picogreen in a control cell and a Mic60 RNAi cell. f Time-lapse images of Mic60 RNAi cells expressing mito-DsRed and stained by picogreen. White arrowheads indicate MDT processes. Orange arrowheads mark the sites of mtDNA. g Percentage of MDT events that drive mtDNA transportation to total MDT events in control and in Mic60 RNAi cells over a 5-min course (n = 10 cells were used per condition from three independent experiments). h KIF5B−/− NRK cells expressing mito-YFP, TFAM-mCherry, and TET-ON-KIF5B contain control or Mic60 siRNA. Both control and Mic60 RNAi cells were treated with 0.5 µg/ml tetracycline for 4 h. Yellow dashed lines indicate the boundary of the mitochondrial network. Red dashed lines indicate the boundary of the perinuclear mitochondria. Inset regions are magnified from the boxed areas. i Western blots of Mic60 indicate depletion of Mic60 in lysates of cells transfected with siRNA against Mic60. j Quantification of nucleoids density in the perinuclear zone and peripheral zone of control or Mic60 RNAi cells after adding 0.5 µg/ml tetracycline for 4 h (n = 20 cells were used per condition from three independent experiments). Scale bars: a, h, 10 μm; inset in a, 5 μm; b–d, f and inset in h, 2 μm; e, 1 μm. Data are presented as mean ± SEM. P value from top to bottom and left to right: p < 0.0001, p = 0.7101, p = 0.0006. NS no significant difference; ***p < 0.001, two-tailed, unpaired Student’s t-test. Source data are provided as a Source data file.