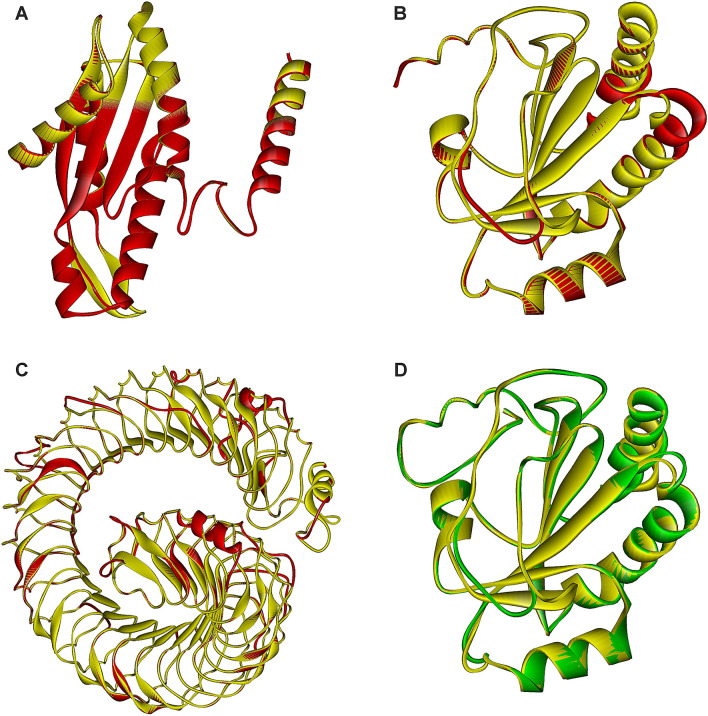
Fig. 3.
Structural models of terminal extra domains of family GH126 members. a The model of the C-terminal extension of the protein from Bacillus velezensis (GenBank accession No.: QHK13041.1; residues S458-E636; red) overlapped with the corresponding part of a signalling protein from Caulobacter vibrioides (PDB code: 1W25; residues L261-K442; yellow); b the model of the N-terminal extension of the protein from Clostridium butyricum (AXB84457.1; residues I41-S207; red) overlapped with the thioredoxin-like fold present in the protein Rv2874 from Mycobacterium tuberculosis (2HYX; residues I376-K545; yellow); c the model of the N-terminal extension of the protein from Lactobacillus brevis (AYM02277.1; residues S37-G759; red) with the leucine-rich-repeat domain present in the Ser/Thr-protein kinase from Arabidopsis thaliana (6S6Q; residues T29-N-859; yellow); and d the model of the N-terminal extension of the protein from Bacteroides xylanolyticus (WP_104434259.1; residues N37-E209; green) overlapped with the thioredoxin-like fold present in the protein Rv2874 from Mycobacterium tuberculosis (2HYX; residues E366-N542; yellow). The individual superimposed parts cover: a 179 Cα-atoms with a 0.24 Å RMSD; b 162 Cα-atoms with a 0.50 Å RMSD; c 676 Cα-atoms with a 0.59 Å RMSD; and d 170 Cα-atoms with a 0.57 Å RMSD. Note, all templates are in each case coloured yellow, whereas the models are shown in red (a, b and c) or green (d) depending on the fact whether or not the protein has already been classified in the family GH126