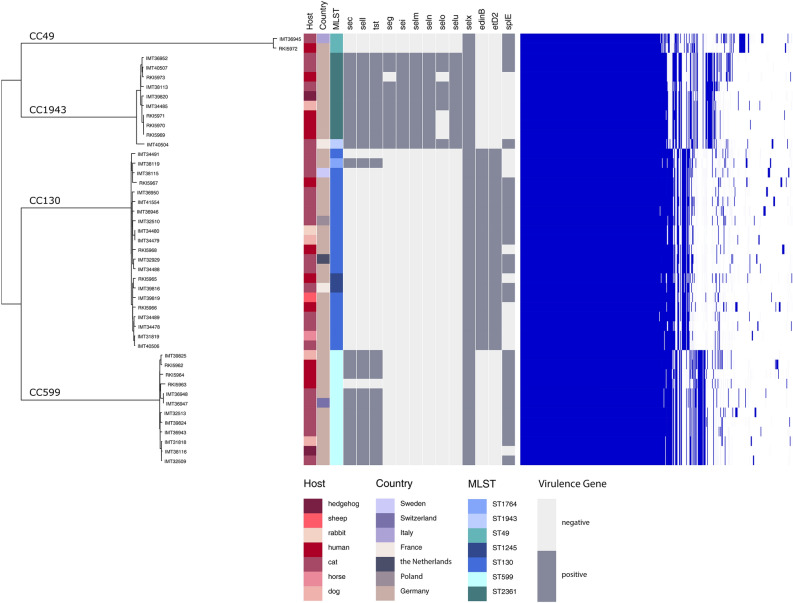
Figure 1.
The core genome phylogeny based on the Maximum Common Genome. The core genome phylogeny based on the Maximum Common Genome comprising 2,094 orthologous genes present in all isolates show four distinct clusters, whereby the genetic diversity within the clusters is rather low. Furthermore, the isolates metadata show no significant association with the core genome clusters. The 2,003 accessory genes show a distribution pattern that is highly correlated with the core genome clusters (right side), suggesting a lineage-specific gene content. Genes for aureolysin (aur), leucotoxins D and E (lukD, lukE), gamma-haemolysin component A–C (hlgA, hlgB, hlgC) and proteases SpIA or SpIB are present in all isolates. All isolates belonging to ST-1943 as well as some CC130 and CC599 were positive for different variants of the Staphylococcal pathogenicity island (SaPI) harbouring a toxic shock toxin encoding gene (tst), which were variants of tst-bov80. Moreover, 48.5% of the mecC-positive isolates harboured staphylococcal enterotoxins (SE). The protease SpIE can just be found in 23/33 isolates and is not associated with any sequence type. The epidermal cell differentiation inhibitor B (edinB) cannot be determined in the isolates of ST-1943, ST-2361, ST-49 and ST-599.