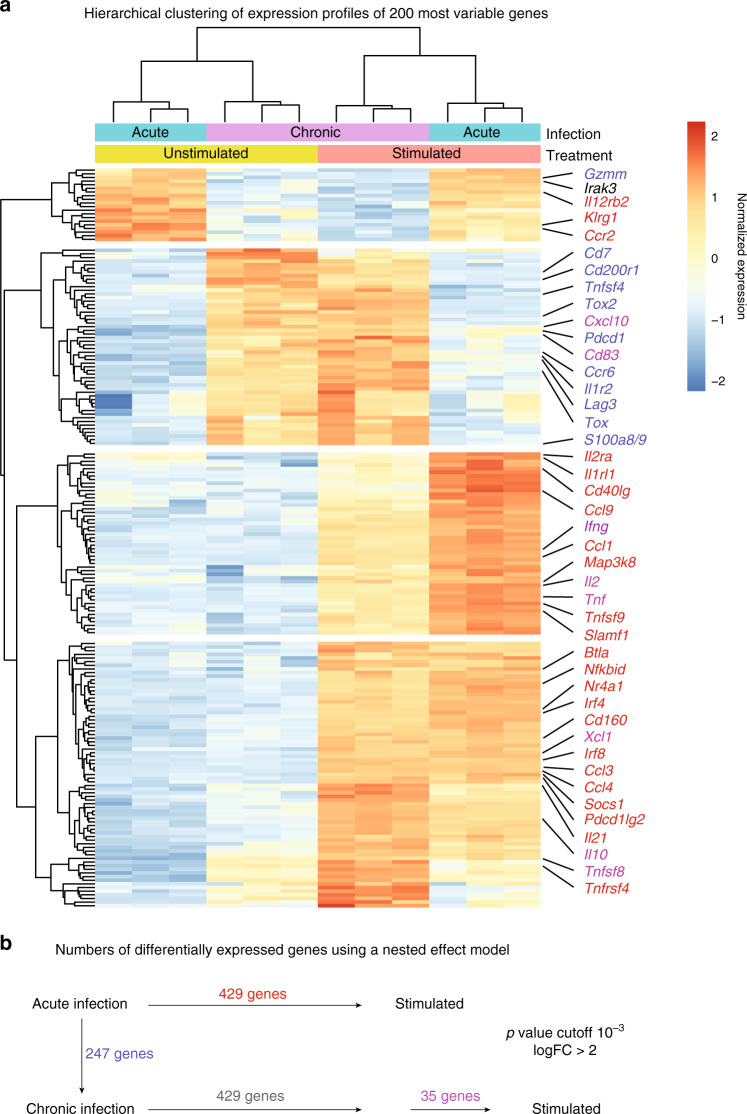
Fig. 1. Transcriptional profiling of functional or exhausted P14 cells with or without restimulation.
P14 cells were adoptively transferred into mice 1 day prior high or low-dose LCMV clone 13 infection. Animals were sacrificed after 14 days. CD8+ P14 cells were stimulated with anti-CD3 and anti-CD28 for 4 h. RNA was extracted and sequenced. a Heatmap of the 200 most variable gene profiles was generated using hierarchical clustering (k = 4). Selected genes are annotated. Heatmap colors indicate normalized gene expression ranging from high (red) to low (blue). Gene color highlights differentially expressed genes between groups indicated in b. b Number of differentially expressed genes between the conditions are indicated. The nested effect model disentangles the effect of chronic infection and stimulation and contains an interaction term for stimulation in chronic infection. Color code of gene names indicates the group of the differential expression analysis (see Methods for details). Data from one experiment are shown. Source data are provided as a Source Data File.