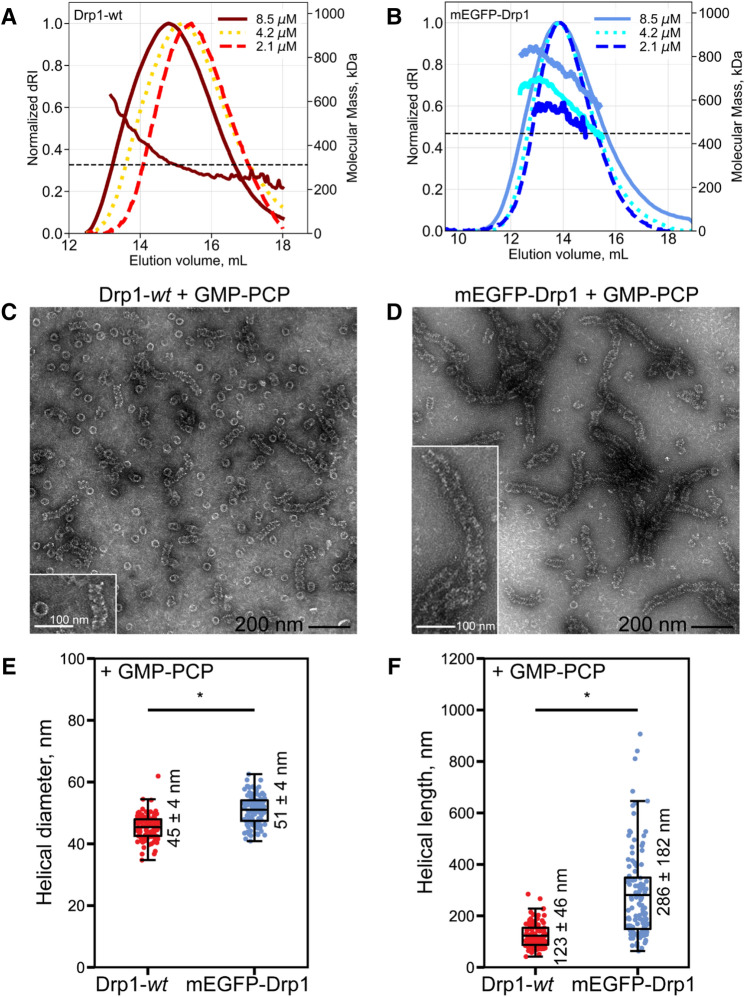
Figure 3.
SEC-MALS and EM reveal altered quaternary structure and higher-order oligomerization states for mEGFP-Drp1 in vitro. (A,B) SEC-MALS profiles of Drp1-wt (A) and mEGFP-Drp1 (B) as a function of total loaded protein concentration before sieving. The horizontal dashed line in each plot indicates the approximate molecular mass of a corresponding tetramer (~ 328 kDa for Drp1-wt and ~ 444 kDa for mEGFP-Drp1). For Drp1-wt, molar mass distribution for only 8.5 μM loading protein concentration is shown. Drp1-wt displayed elution profile shifts as a function of protein concentration typically observed for self-associating systems in dynamic equilibrium. mEGFP-Drp1 showed no such apparent elution profile shifts and primarily sampled tetramer or higher oligomeric states. (C,D) Representative negative-stain EM images of Drp1-wt (C) and mEGFP-Drp1 (D) in the presence of GMP-PCP. Scale bar, 200 nm. Insets show magnified images of representative self-assembled higher-order structures. Inset scale bar, 100 nm. (E,F) Boxplots of helical diameter (E) and lengths (F) comparing Drp1-wt and mEGFP-Drp1 in the presence of GMP-PCP. Statistically significant differences between groups are indicated by a star. Mean ± S.D. is indicated next to each box. For helical diameters in panel E, Drp1-wt n = 114; mEGFP-Drp1 n = 105, p < 0.0001. For helical lengths in panel F, Drp1-wt n = 116; mEGFP-Drp1 n = 117, p < 0.0001. SEC-MALS profiles and box and whisker plots were prepared using matplotlib v3.2.2 available at https://matplotlib.org/3.2.2/api/_as_gen/matplotlib.pyplot.boxplot.html.