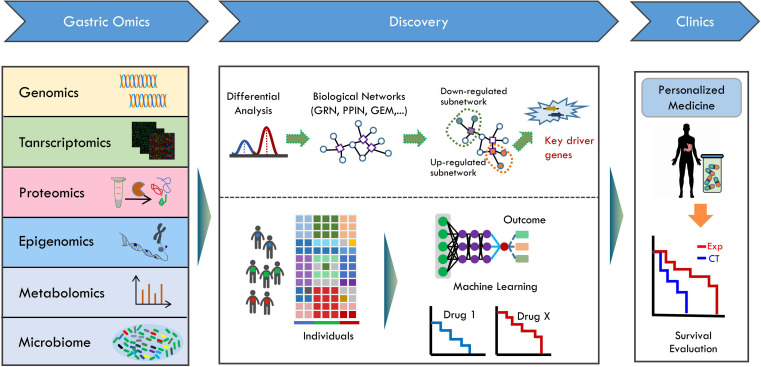
FIGURE 1.
The systems biology approach for gastric cancer research. The different types of omics data including genomics, transcriptomics, proteomics, epigenomics, and metabolomics are obtained using according omics technologies from gastric cancer cohort. Omics data (transcriptome, proteome, or metabolome) is measured for two or more group that differ in clinical information. Generally, differentially expressed genes (DEGs) based methodology is applied. DEGs are identified by comparative analysis of measured omics data. With integrative network-based approach, the biological networks, such as gene regulatory network (GRN), protein-protein interaction network (PPIN), and genome-scale metabolic network (GEM) are used together with omics data in an integrative way. Then, the up-regulated or down-regulated subnetworks are identified by integrating data into network models. Using network modeling tools, the key driver gene linked to clinical information can be identified. Identified key driver genes can be further applied into clinical studies. In addition, the multi-omics data are used to stratify patient into subtypes. Machine learning algorithms utilize omics features to predict potential treatment outcome. The machine learning methodology can be applied to chemotherapy, immunotherapy or their combinations. Finally, the key driver gene information and the predictive models can be used to design the personalized treatment strategy, and applied in clinics. The clinical survival outcome then can be evaluated after personalized medicine treatment.