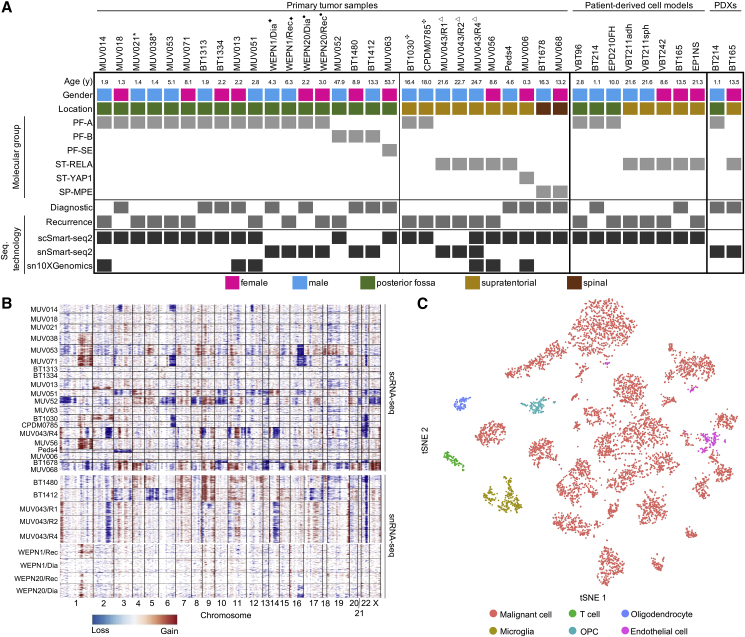
Figure 1.
Classification of Human EPN Single-Cell Transcriptomes
(A) Clinical and molecular details of the human EPN dataset of fresh/frozen patient samples (n = 28), patient-derived cell models (n = 8), and PDXs (n = 2). scRNA-seq technologies applied per sample are indicated. Matched pairs (n = 5) are indicated by superscript symbols.
(B) Inference of copy-number alterations (CNAs) from scRNA-seq (top) and snRNA-seq (bottom) data on the basis of average relative expression of sliding windows of 100 genes. Each row corresponds to a cell, ordered by tumor and clustered within each tumor by CNA patterns.
(C) t-Distributed stochastic neighbor embedding (tSNE) of all cells derived from scRNA-seq and snRNA-seq. Cells are colored according to presence of CNAs and similarity to expression signatures of non-malignant cell populations (T cells, oligodendroglial precursor cells [OPC], oligodendrocytes, microglia, endothelial cells).
See also Table S1.