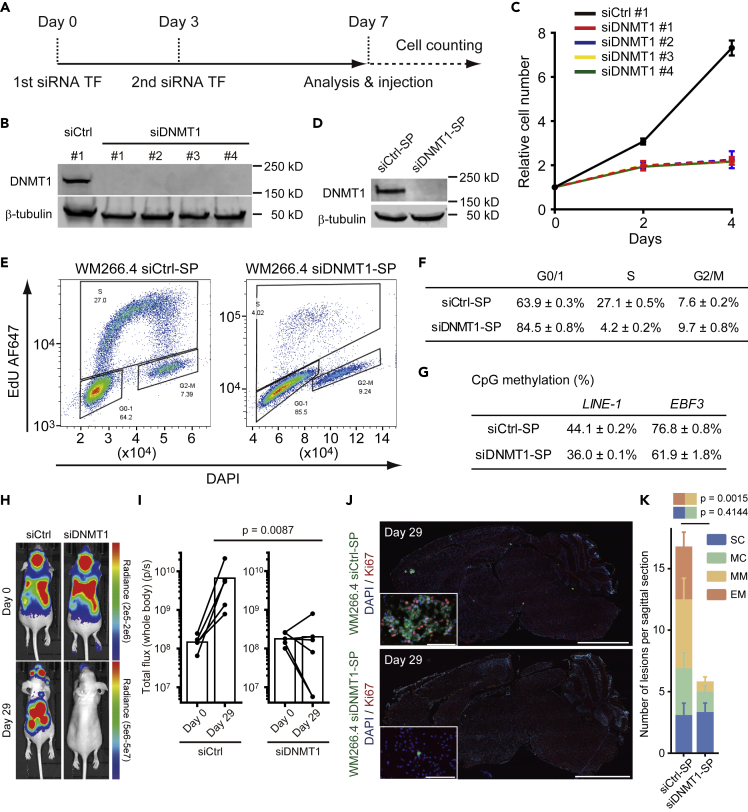
Figure 3.
DNMT1 Suppression Induces Cell Cycle Delay in WM266.4 Cells
(A) The time course of siRNA experiments. WM266.4 cells were transfected with control siRNA or siRNA against DNMT1 at days 0 and 3, and further experiments were started at day 7.
(B) Immunoblotting at day 7 showed effective ablation of DNMT1 in WM266.4 cells treated with four different siRNAs (siDNMT1 #1–4).
(C) Cell proliferation analysis of WM266.4 cells treated with siCtrl (#1) or siDNMT1 (#1–4).
(D) Immunoblotting at day 7 showed effective ablation of DNMT1 in WM266.4 cells treated with siCtrl smart pool (siCtrl-SP) or siDNMT1 smart pool (siDNMT1-SP).
(E and F) Cell cycle analysis of WM266.4 cells treated with siCtrl-SP or siDNMT1-SP. Representative FACS images with cell cycle profiling by EdU incorporation (y-axis) and DNA content (x-axis, DAPI staining) are shown in (E) and the percentages of the cells in G0/1, S and G2/M phase are shown in (F) (n = 3).
(G) CpG methylation analysis at transcription start site of the indicated genes by pyrosequence. LINE-1 is used as a marker of global DNA methylation (average of three CpG sites, n = 2) and EBF3 as a representative highly methylated region in melanoma cells (n = 2).
(H and I) WM266.4 cells pre-treated with siCtrl-SP or siDNMT1-SP were injected into mouse hearts and induced brain metastasis. Representative images of bioluminescence detection (H) and total flux from the whole body (photons/s) at day 0 (after injection) and day 29 (siCtrl-SP; n = 5, siDNMT1-SP; n = 6) (I) are shown.
(J and K) Representative images of sagittal brain sections stained for Ki67 (J) and the number of lesions per sagittal brain section at day 29 (K) are shown. A total of 10 brain slices from 5 mice (siCtrl-SP) and 12 brain slices from 6 mice (siDNMT1-SP) were used for quantification. Green: WM266.4 cells pre-treated with siCtrl-SP or siDNMT1-SP, Blue: DAPI, Red: Ki67. Scale bar, 2.5 mm (large images), 100 μm (small panels).
Data are mean (I), mean ± SD (C, F, and G), or mean ± SEM (K).