Figure 5.
Brain Microenvironment Activates Pro-survival Signals via DNMT1 Suppression
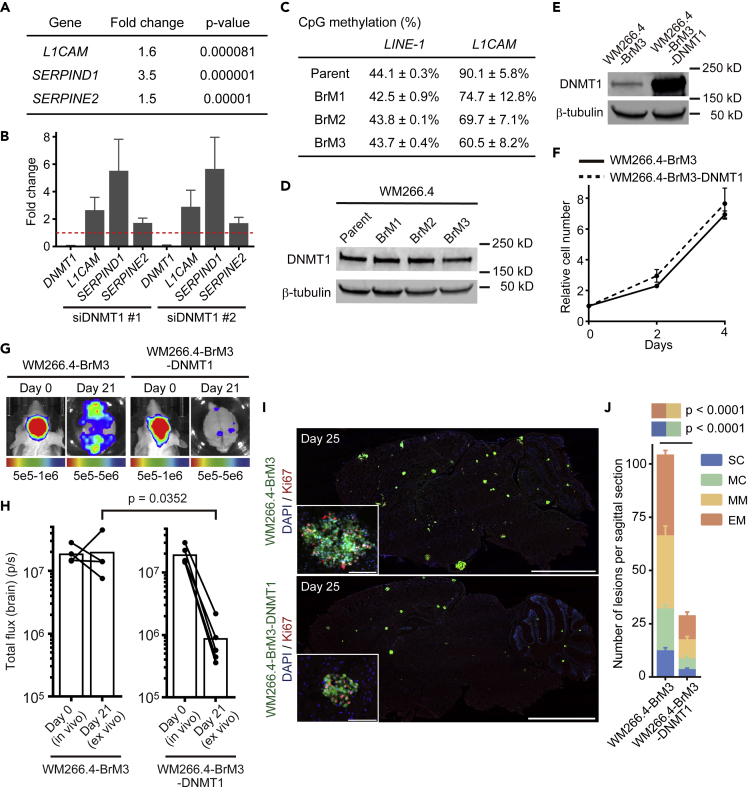
(A) Transcriptome analysis of WM266.4 cells treated with siCtrl-SP or siDNMT1-SP. Genes up-regulated by DNMT1 knockdown are shown with fold change and p value (n = 3). Please also refer to Data S6.
(B) Relative expression of the indicated genes in WM266.4 cells treated with siDNMT1-#1 or -#2 compared with siCtrl-#1 were quantified by quantitative RT-PCR.
(C) CpG methylation analysis of the indicated genes in WM266.4 (parent) and its derivatives (BrM1-3) by pyrosequence. The methylation of L1CAM is the average of five CpG sites in the promoter region (n = 2), and LINE-1 is used as a marker of global DNA methylation (average of three CpG sites, n = 2).
(D) Immunoblotting for DNMT1 in WM266.4 (parent) and its derivatives with enhanced brain metastasis (BrM1-3).
(E) Immunoblotting for DNMT1 in WM266.4-BrM3 cells and WM266.4-BrM3 cells overexpressing DNMT1 (WM266.4-BrM3-DNMT1).
(F) Cell proliferation analysis of WM266.4-BrM3 and WM266.4-BrM3-DNMT1 cells (n = 3).
(G and H) WM266.4-BrM3 or WM266.4-BrM3-DNMT1 cells were injected into mouse hearts and induced brain metastasis. Representative images of bioluminescence detection in vivo (day 0) and ex vivo (day 21) (G) and total flux from the brain (photons/s) (WM266.4-BrM3; n = 4, WM266.4-BrM3-DNMT1; n = 5) (H) are shown.
(I and J) Representative images of sagittal brain sections stained for Ki67 (I) and the number of lesions per sagittal brain section at day 25 (J) are shown. A total of 12 brain slices from 6 mice were used for quantification in each group. Green: WM266.4-BrM3 or WM266.4-BrM3-DNMT1 cells, Blue: DAPI, Red: Ki67. Scale bar, 2.5 mm (large images), 100 μm (small panels).
Data are mean (H), mean ± SD (B, C, and F), or mean ± SEM (J).