Figure 8.
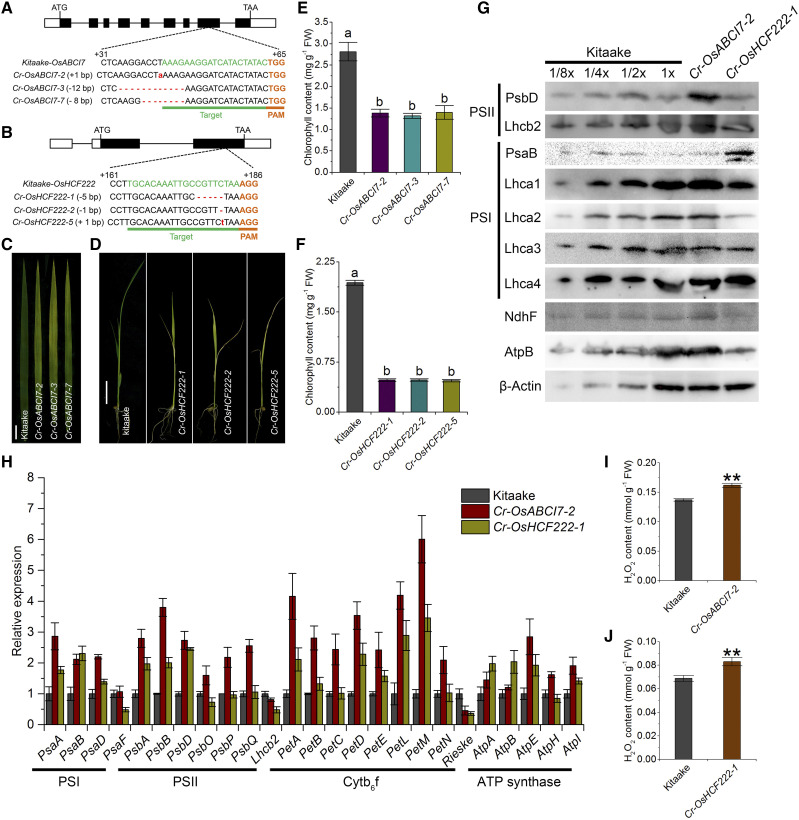
Characterization of CRISPR/Cas9-edited OsABCI7 and OsHCF222 mutations in the Kitaake background. A and B, CRISPR/Cas9-mediated mutations at the target sites of OsABCI7 and OsHCF222 in representative knockout lines. The sgRNA target sequence is underlined in green, and the PAM motif is indicated in orange. C, Phenotypes of flag leaves in Kitaake and Cr-OsABCI7 knockout lines at 60 d after transplanting. D, Seedlings of T1 Cr-OsHCF222 knockout lines at 12 d after germination. Bars = 2 cm. E and F, Chlorophyll contents of Cr-OsABCI7 and Cr-OsHCF222 knockout lines in C and D. Different letters indicate significant differences by one-way ANOVA and Duncan’s test (P < 0.05). Data are means ± sd (n = 3). G, Levels of thylakoid membrane proteins tested in total proteins extracted from Kitaake, Cr-OsABCI7, and Cr-OsHCF222 knockout lines at 8 d after germination. H, Detection of mRNA levels of plastid- and nucleus-encoded genes related to thylakoid membrane proteins in Cr-OsABCI7-2 and Cr-OsHCF222-1 lines at 8 DAG. I and J, Measurements of H2O2 contents in Cr-OsABCI7-2 and Cr-OsHCF222-1 at 8 DAG. Data are means ± sd (n = 3). Asterisks indicate significant difference by Student’s t test (**P < 0.01). FW, Fresh weight.