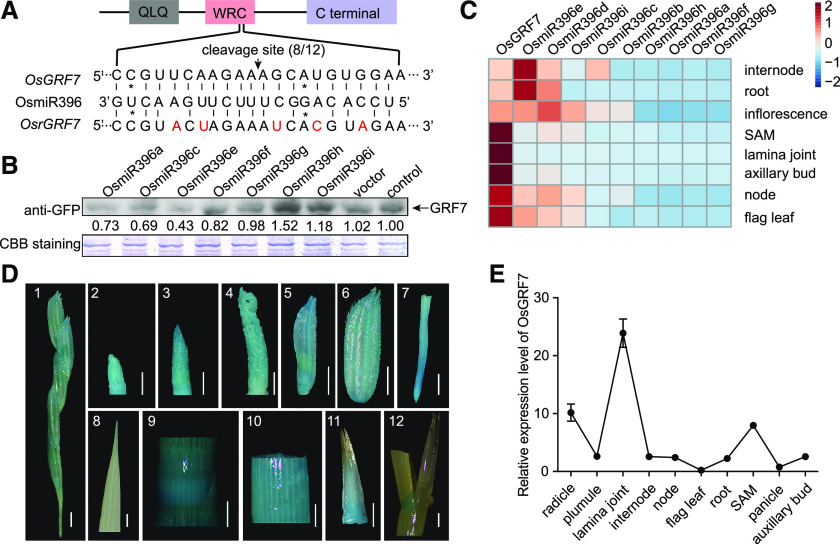
Figure 2.
OsGRF7 is mainly repressed by OsmiR396e and expressed in various tissues. A, The OsGRF7 cleavage site sequence complementary to OsmiR396. The position corresponding to the 5′ end of the cleaved OsGRF7 mRNA determined by 5′ RACE (rapid amplification of cDNA end) and the frequency of 5′ RACE clones corresponding to the cleavage site is shown by the arrow. 8/12 means eight of 12 clones have an OsmiR396 cleavage site. The mutated sites in OsrGRF7 are marked in red. The asterisks indicate mismatched sites between OsmiR396 and OsGRF7. The gray box indicates the QLQ domain, the pink box indicates the WRC domain, and the purple box indicates the C-terminal end of OsGRF7. B, Protein immunoblotting of OsGRF7 coexpressed with OsmiR396s in GRF7-GFP transgenic rice protoplasts. Vector and control mean with or without the empty vector, respectively. Coomassie Brilliant Blue (CBB) staining was used as a loading control. The relative protein amounts were determined by ImageJ (National Institutes of Health). C, Relative expression levels of OsGRF7 and OsmiR396s were detected by RT-qPCR in internode, root, inflorescence (10 cm), node, flag leaf, shoot apical meristem (SAM), axillary bud, and lamina joint. D, GRF7pro:GUS expression patterns in transgenic plants. Images are as follows: 1, small panicle branch; 2 to 4, inflorescence at different stages; 5 and 6, spikelet; 7, root tip; 8, flag leaf; 9, node; 10, internode; 11, axillary bud; 12, lamina joint of the 30-d seedling. Bars = 2 mm. E, Relative expression levels of OsGRF7 in different tissues analyzed with RT-qPCR. OsUBI was used as an internal reference. Values are means ± sd of three biological replicates.