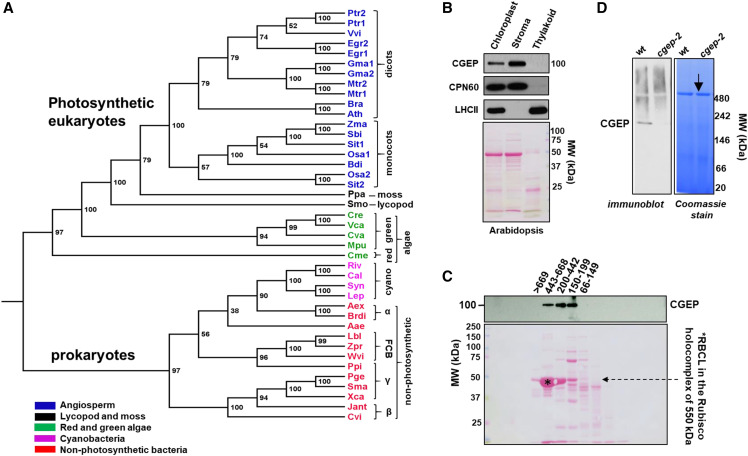
Figure 1.
Phylogenetic analysis and subcellular localization of CGEP A, Phylogenetic tree of 41 CGEP homologs showing two distinct clades: one with photosynthetic eukaryotes (mono- and dicotyledons, moss, a lycopod, and red and green algae) and one with only photosynthetic and nonphotosynthetic prokaryotes. Within these prokaryotes, CGEPs are found in many cyanobacteria (marked in pink) and many Gram-negative bacteria (marked in red; proteo- and flavo-bacteria). CGEP homologs were not obvious in other major branches of the tree-of-life (e.g. archaea, fungi, animals). Bootstrap values are shown. A complete list with the explanations of the abbreviations is provided in Supplemental Table S1 and complete alignment is given in Supplemental Dataset S1. B, CGEP localized to Arabidopsis stroma determined by immunoblotting. CPN60 and LHCII were used as marker proteins for stroma and thylakoid, respectively. Ponceau-stained blot (before immunodetection) is shown below. C, Gel filtration of Arabidopsis stromal proteome showing that CGEP accumulates in complexes between ∼150 and 600 kD. The Rubisco holocomplex at 550 kD is marked by an asterisk. D, Native PAGE of Arabidopsis chloroplast stroma shows CGEP dimerization in wild-type plants. The arrow indicates the Rubisco holocomplex at 550 kD. cgep-2 refers to an Arabidopsis null mutant for CGEP.