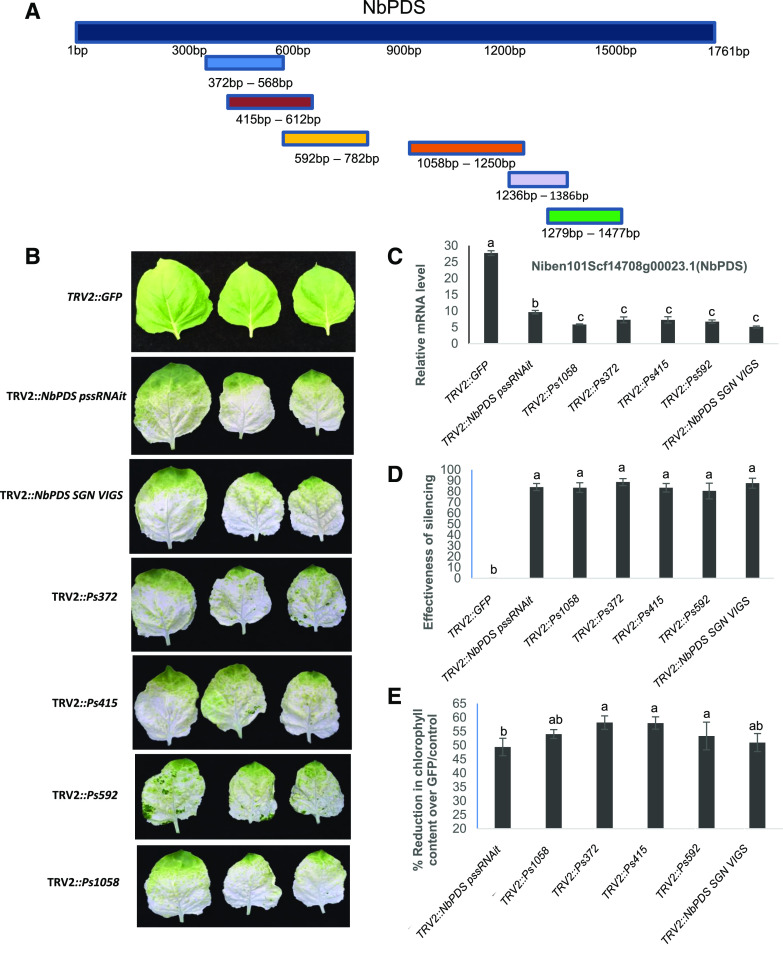
Figure 6.
VIGS constructs and their effectiveness in silencing the PDS gene in N. benthamiana. A, Schematic representation of the VIGS fragments located on the NbPDS gene, based on the webserver prediction. Ps series VIGS fragments and TRV2::NbPDS pssRNAit fragments were predicted using pssRNAit. The TRV2::NbPDS SGN VIGS fragment was designed using the VIGS SGN tool. B, Expression analysis showing silencing of NbPDS in N. benthamiana VIGS plants. C, Phenotype of the silenced plants. The top three leaves were photographed and assessed for effectiveness. D, Effectiveness of the silencing as assessed by the number of leaves that turned white versus the number of green leaves. E, Percent reduction in chlorophyll content in silenced plants compared to the GFP fragment as a control. Average values of three biological replicates were used to generate bar graphs and experiments were repeated three times with similar results. Error bars indicate the se. Different letters above the bars indicate a significant difference from two-way ANOVA at P < 0.05 with Tukey’s HSD mean-separation test (α = 0.05).