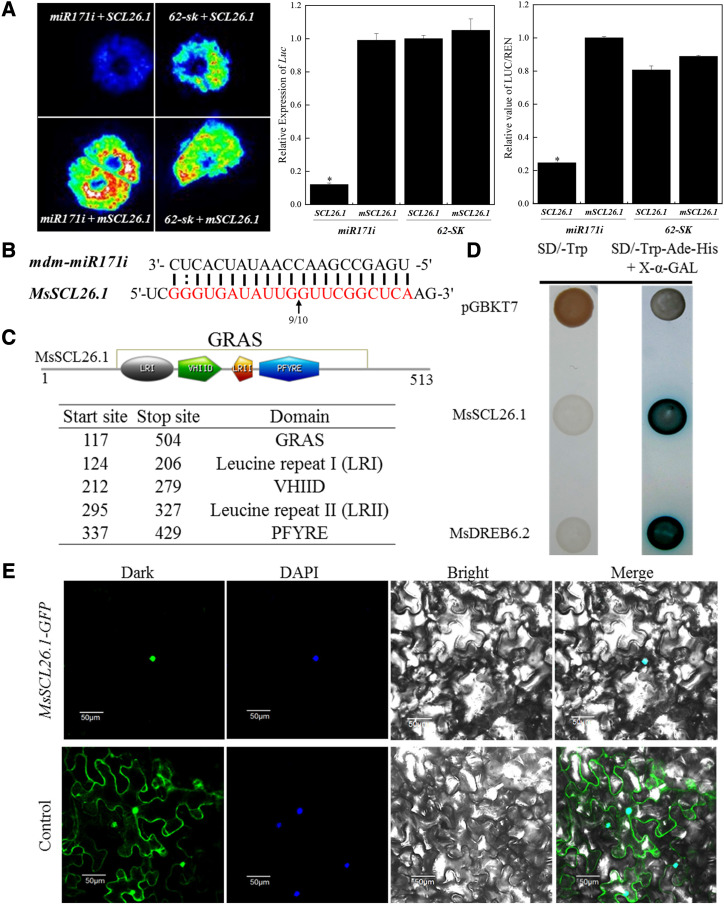
Figure 3.
The interactional mode between mdm-miR171i and MsSCL26.1 and characterization of the GRAS transcription factor MsSCL26.1. A, Dual LUC reporter assay to qualitatively and quantitatively evaluate mdm-miR171i-mediated repression of the target gene MsSCL26.1. (MsCL26.1 is a synonymous mutation of MsSCL26.1 in mdm-miR171i cleavage sites.) Renilla luciferase was used as a positive control in the pGreen-dualluc-ORF-sensor vector. Data are presented as means ± sd from three biological replicates. Asterisks indicate significant differences between the test group and control group based on Duncan’s multiple range test (*P < 0.05). B, mdm-miR171i-directed cleavage site in MsSCL26.1 identified by 5′-RACE. The arrow indicates the mdm-miR171i cleavage site in the MsSCL26.1 transcript. The numbers represent the frequency of clones used to validate the cleavage sites of the target mRNAs. C, Four typical GRAS protein domains in MsSCL26.1. D, Transactivation activity assay of MsSCL26.1 in yeast strain AH109. MsDREB6.2 was used as a positive control. E, Subcellular localization of MsSCL26.1. GFP (Control) and the MsSCL26.1-GFP fusion gene driven by the CaMV 35S promoter were separately transiently expressed in N. benthamiana leaf epidermal cells. DAPI (4’,6-diamidino-2-phenylindole) was used as a nuclear marker.