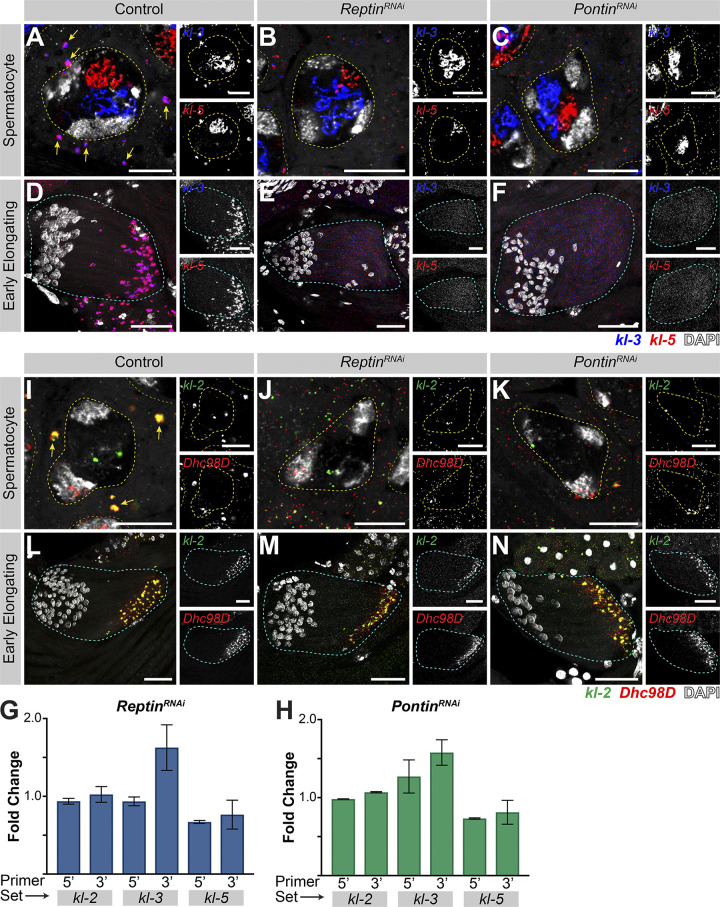
Figure 4.
Rept and Pont are required for kl-granule assembly. (A–F) smFISH against kl-3 and kl-5 in control (A and D), rept RNAi (bam-gal4>UAS-reptKK105732; B and E), or pont RNAi (bam-gal4>UAS-pontKK101103; C and F) SCs (A–C, single z plane) and early elongating spermatids (D–F, z-projection). Shown are kl-3 (blue), kl-5 (red), DAPI (white), SC nuclei (yellow dashed lines), neighboring SC nuclei (narrow yellow dashed lines), SC kl-granules (yellow arrows), and a spermatid cyst (cyan dashed line). Scale bars: 10 µm (A–C) or 25 µm (D–F). (G and H) Quantitative RT-PCR in rept RNAi (bam-gal4>UAS-reptKK105732, G) or pont RNAi (bam-gal4>UAS-pontKK101103, H) for kl-3, kl-5, and kl-2 using two primer sets per gene, as indicated (see Table S1). Data were normalized to GAPDH and sibling controls and represent at least two biological replicates, each reaction performed in technical triplicate. Error bars represent SD. (I–N) smFISH against kl-2 and Dhc98D in control (I and L), rept RNAi (bam-gal4>UAS-reptKK105732, J and M), or pont RNAi (bam-gal4>UAS-pontKK101103; K and N) SCs (I–K, single z plane) and early elongating spermatids (L–N, z-projection). Shown are kl-2 (green), Dhc98D (red), DAPI (white), SC nuclei (yellow dashed lines), neighboring SC nuclei (narrow yellow dashed lines), SC kl-granules (yellow arrows), and a spermatid cyst (cyan dashed line). Scale bars: 10 µm (I–K) or 25 µm (L–N).