Figure S1.
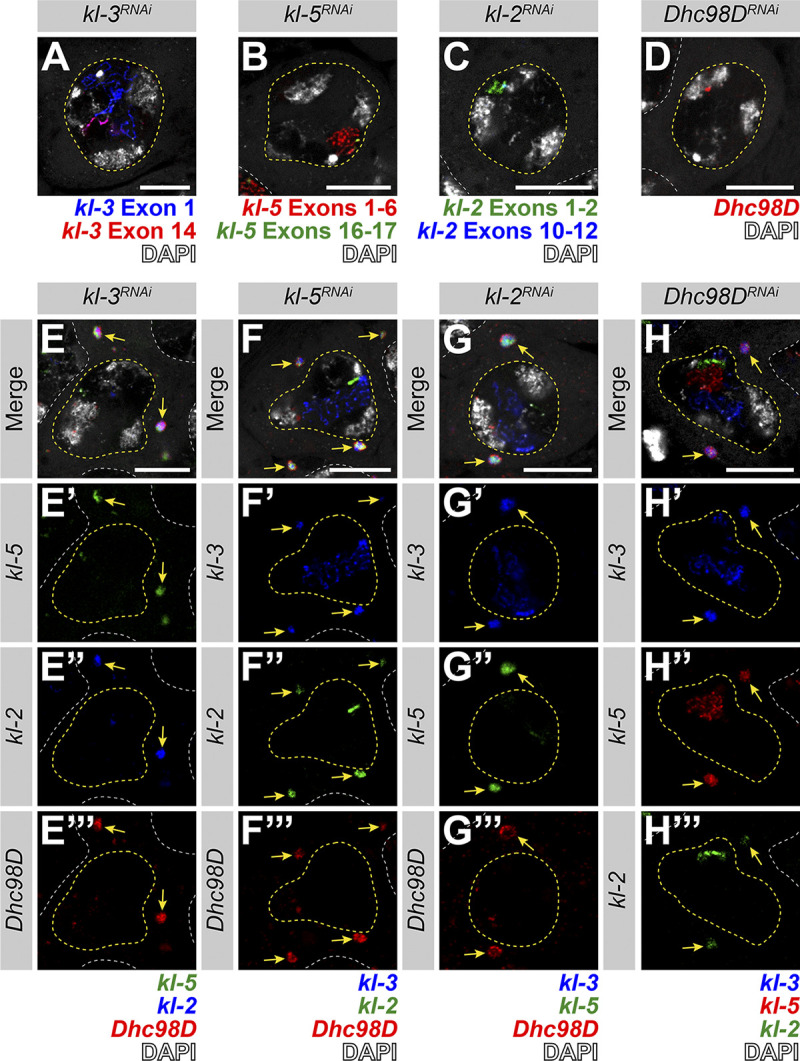
kl-granule formation is not dependent upon any one mRNA constituent. (A–D) smFISH against each known kl-granule mRNA constituent following RNAi of that constituent shows successful knockdown (no remaining cytoplasmic signal). Note that we use multiple smFISH probe sets for some mRNAs targeted against different regions of the transcript (see Table S1). (A) kl-3 exon 1 (blue), kl-3 exon 14 (red) and DAPI (white). (B) kl-5 exons 1–6 (red), kl-5 exons 16–17 (green), and DAPI (white). (C) kl-2 exons 1–2 (green), kl-2 exons 10–12 (blue), and DAPI (white). (D) Dhc98D (red) and DAPI (white). For all, SC nuclei, yellow dashed line; neighboring SC nuclei, white dashed line. Scale bars: 10 µm. (E–H) smFISH against the other three constituent mRNAs after RNAi of the fourth mRNA. Note that the color used to represent each smFISH probe corresponds to the probe sets in A–D. (E) kl-5 (green), kl-2 (blue), Dhc98D (red) and DAPI (white). (F) kl-3 (blue), kl-5 (green), Dhc98D (red), and DAPI (white). (G) kl-3 (blue), kl-5 (green), Dhc98D (red) and DAPI (white). (H) kl-3 (blue), kl-5 (red), kl-2 (green), and DAPI (white). For all, SC nuclei, yellow dashed line; neighboring SC nuclei, white dashed line; kl-granules, yellow arrows. Scale bars: 10 µm.
