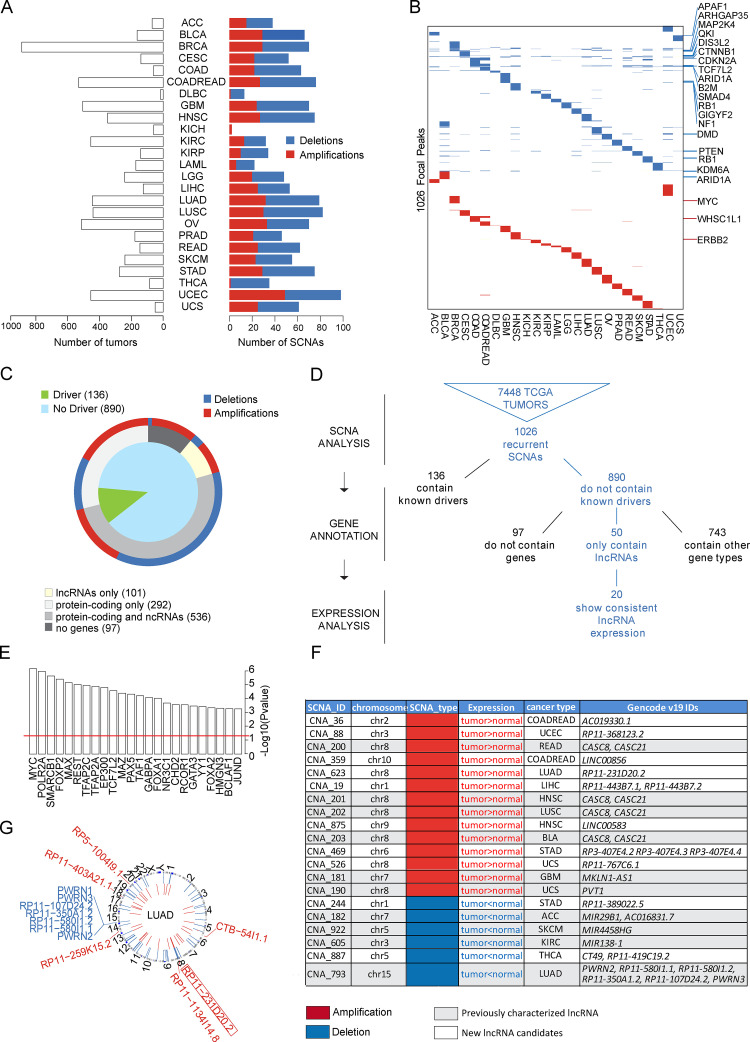
Figure 1.
Overview of cancer-associated SCNAs that contain lncRNAs. (A) Distribution of the TCGA tumor samples and SCNAs detected in the 25 types of cancer. (B) Distribution of SCNAs in the different cancer types, indicating some of the high-confidence cancer drivers (Tamborero et al., 2013) within the alterations. (C) Classification of the SCNAs based on the biotypes of the genes inside them. (D) Pipeline for the selection of SCNAs harboring lncRNAs, integrating SCNA detection, gene annotation, and expression analysis. The starting point of the analysis is the copy number data from 7,448 tumors, in which 1,055 recurrent SCNAs were detected. Then these SCNAs were classified based on gene annotation and gene expression analysis. (E) Transcription factors with binding significantly enriched around the TSS of amplified lncRNAs. Statistical significance was determined by hypergeometric test; P values were −log10 transformed. (F) SCNAs selected by the analysis as containing putative cancer-relevant lncRNAs. (G) Circos plot indicating the copy number–altered lncRNAs present in lung adenocarcinomas. Indicated in bold and squared is the lncRNA RP11-231D20.2 selected for the present study. ncRNA, noncoding RNA; ACC, adrnocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; COAD, colon adenocarcinoma; COADREAD, colorectal adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; OV, ovarian serous cystadenocarcinoma; PRAD, prostate adenocarcinoma; READ, rectum adenocardinoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma.