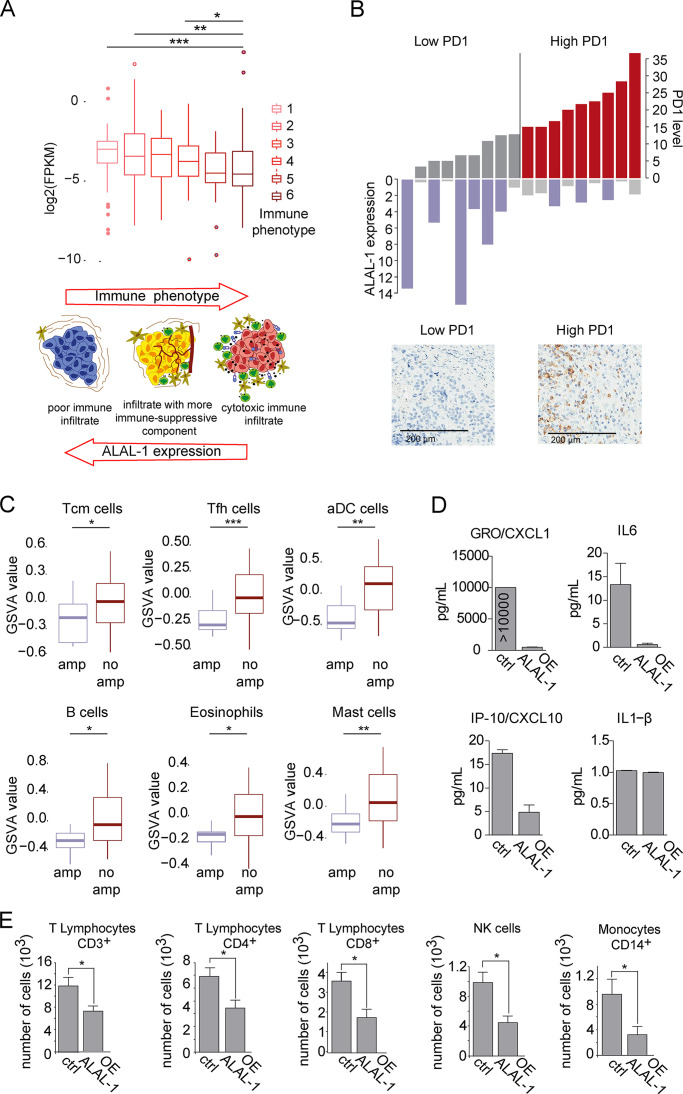
Figure 7.
ALAL-1 contributes to the immune evasion of lung tumors. (A) Expression of ALAL-1 in TCGA-LUSC cohort, classified based on different immune phenotypes based on gene expression signatures of different infiltration patterns and levels of cytotoxic cells (Tamborero et al., 2018). Schematic representation adapted from Tamborero et al. (2018). Significance was determined by two-tailed unpaired Wilcoxon test. (B) ALAL-1 expression and level of PD-1–positive cells in an independent cohort of LUSC tumors. (C) Level of infiltration of the indicated cell populations computed as Gene Set Variation Analysis (GSVA) score in the TCGA-LUAD cohort of tumors with (n = 39) or without (n = 320) ALAL-1 gene amplification. Significance was determined by two-tailed unpaired Wilcoxon test. (D) Level of cytokines secreted to the media of A549 cells that overexpress ALAL-1 asquantified by Luminex assay. (E) Number of cells of different subpopulations from peripheral blood migrated to A549 cells overexpressing ALAL-1 or A549 control cells. Graphs of mean (± SEM) for three independent experiments are shown. Significance was determined by two-tailed unpaired t test and represented as *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. FPKM, fragments per kilobase of exon model per million reads mapped; Tcm, T cell memory; Tfh, T follicular helper cells; aDC, artificial dendritic cells; amp, amplification; GRO, chemokine (C-X-C motif) ligand 1; OE, overexpression; IP-10, C-X-C motif chemokine ligand 10; NK, natural killer; ctrl, control.