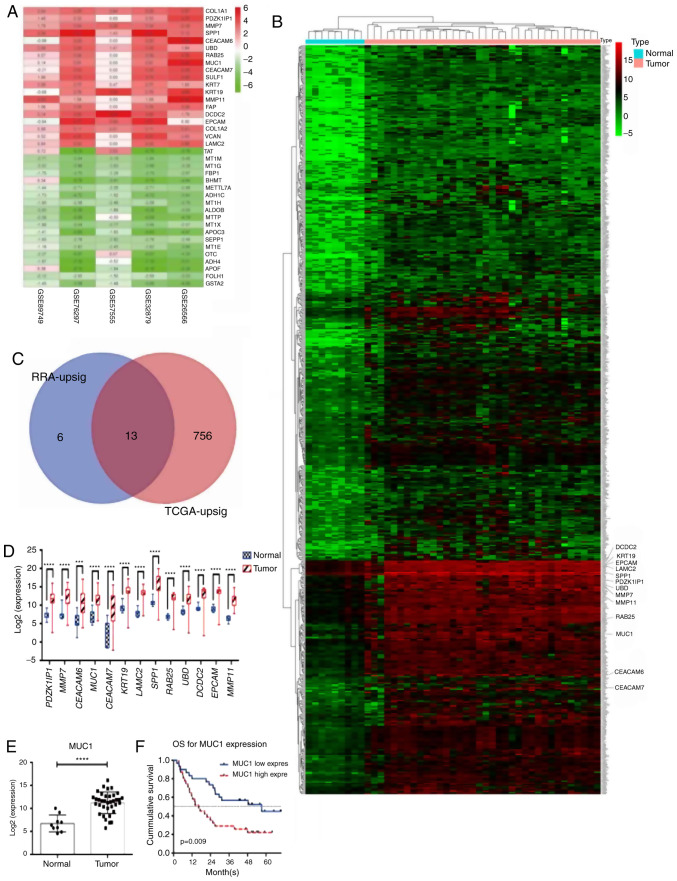
Figure 1.
MUC1 is upregulated in ICC tissue, as validated using databases. (A) The 19 upregulated genes and the 19 most significant downregulated genes linked to cholangiocarcinoma identified using robust rank aggregation. Numbers in the colored fields indicate the log2-transformed fold change. 0.00 or white boxes indicates that the gene was absent from the respective dataset. (B) Upregulated genes linked to cholangiocarcinoma identified using the edgeR package (TCGA dataset). The enlarged gene symbols adjacent to the heatmap are the 13 intersected genes. (C) 13 genes were identified to be upregulated in the RRA integrated datasets and the TCGA data. (D and E) The expression levels of (D) the 13 genes and (E) MUC1 were significantly higher in ICC tissue, validated using TCGA data. (F) Among the 13 genes, only MUC1 predicted OS. RRA-upsig refers to the significantly upregulated genes identified using RRA; TCGA-upsig refers to significantly upregulated genes among the TCGA data. ***P<0.001; ****P<0.0001. MUC1, mucin 1; TCGA, The Cancer Genome Atlas; ICC, intrahepatic cholangiocarcinoma; OS, overall survival; RRA, robust rank aggregation; PDZK1IP1, PDZK1 Interacting Protein 1; MMP7, matrix metalloproteinase 7; CEACAM6, carcinoembryonic antigen related adhesion molecules 6; MUC1, mucin 1; CEACAM7, carcinoembryonic antigen related adhesion molecules 7; KRT19, keratin 19; LAMC2, laminin subunit gamma-2; SPP1, secreted phosphoprotein 1; RAB25, Ras Genes from Brain Protein 25; UBD, ubiquitin D; DCDC2, doublecortin domain containing 2; EPCAM, epithelial cell adhesion molecule; MMP11, matrix metalloproteinase 11.